Zinc »
PDB 8q1y-8qf5 »
8qen »
Zinc in PDB 8qen: Cryo-Em Structure of Apo Clostridioides Difficile Toxin B
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryo-Em Structure of Apo Clostridioides Difficile Toxin B
(pdb code 8qen). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Cryo-Em Structure of Apo Clostridioides Difficile Toxin B, PDB code: 8qen:
In total only one binding site of Zinc was determined in the Cryo-Em Structure of Apo Clostridioides Difficile Toxin B, PDB code: 8qen:
Zinc binding site 1 out of 1 in 8qen
Go back to
Zinc binding site 1 out
of 1 in the Cryo-Em Structure of Apo Clostridioides Difficile Toxin B
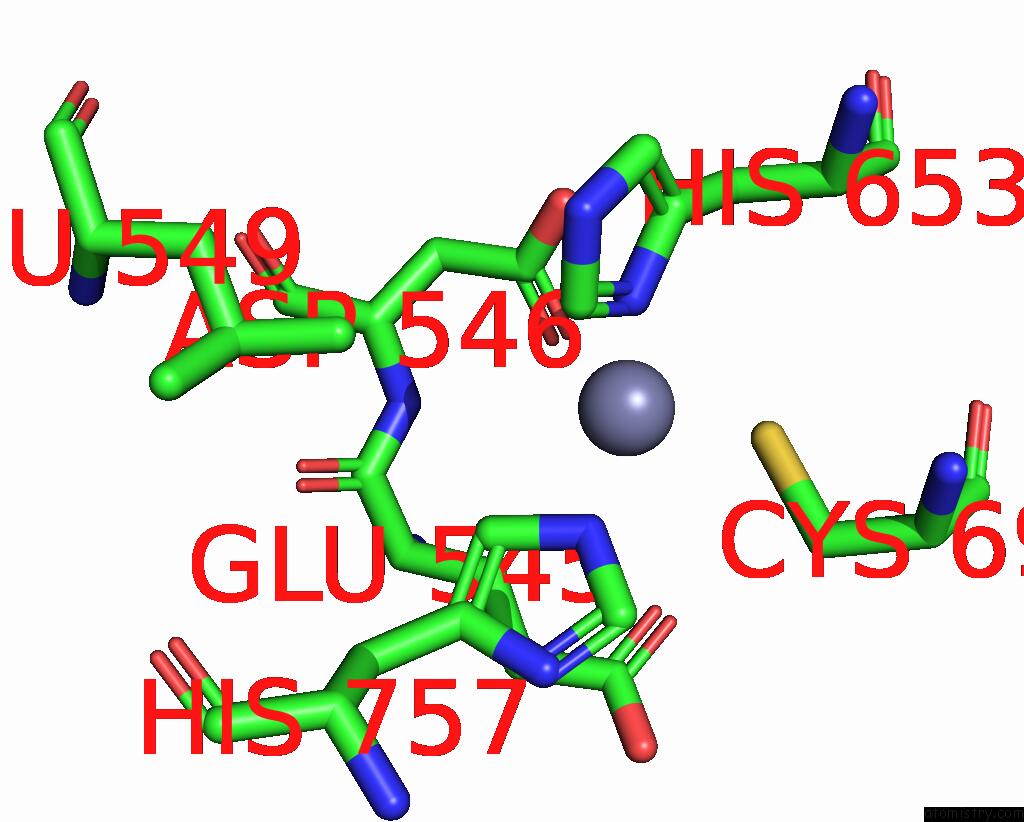
Mono view
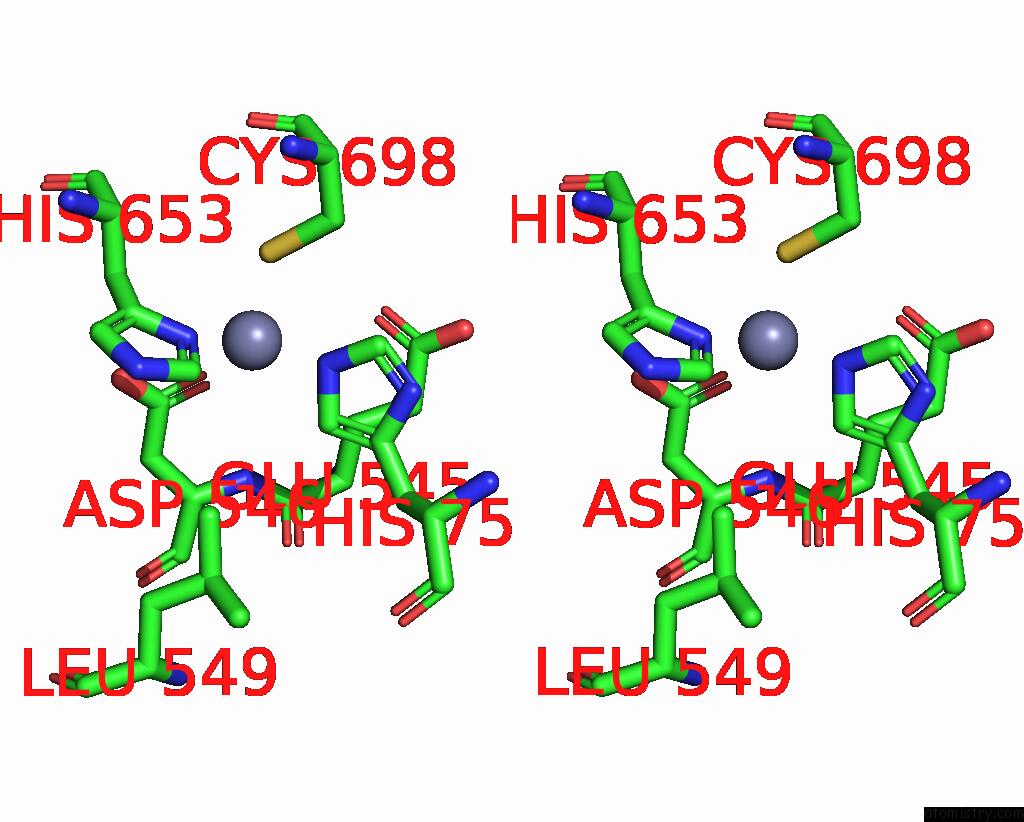
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryo-Em Structure of Apo Clostridioides Difficile Toxin B within 5.0Å range:
|
Reference:
J.Kinsolving,
J.Bous,
P.Kozielewicz,
S.Kosenina,
R.Shekhani,
L.Gratz,
G.Masuyer,
Y.Wang,
P.Stenmark,
M.Dong,
G.Schulte.
Structural and Functional Insight Into the Interaction of Clostridioides Difficile Toxin B and Fzd 7. Cell Rep V. 43 13727 2024.
ISSN: ESSN 2211-1247
PubMed: 38308843
DOI: 10.1016/J.CELREP.2024.113727
Page generated: Fri Aug 22 12:37:21 2025
ISSN: ESSN 2211-1247
PubMed: 38308843
DOI: 10.1016/J.CELREP.2024.113727
Last articles
Zn in 9CP7Zn in 9CNX
Zn in 9CM6
Zn in 9CKF
Zn in 9CKC
Zn in 9CMB
Zn in 9CKG
Zn in 9CLP
Zn in 9CIV
Zn in 9CJA