Zinc »
PDB 8cdb-8cro »
8co4 »
Zinc in PDB 8co4: Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
Enzymatic activity of Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
All present enzymatic activity of Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina:
1.1.1.1; 1.1.1.284;
1.1.1.1; 1.1.1.284;
Protein crystallography data
The structure of Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina, PDB code: 8co4
was solved by
S.Fermani,
S.Fanti,
G.Carloni,
J.Rossi,
G.Falini,
M.Zaffagnini,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 81.92 / 1.90 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 88.6, 93.927, 167.484, 90, 90, 90 |
| R / Rfree (%) | 16.1 / 21.1 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
(pdb code 8co4). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 8 binding sites of Zinc where determined in the Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina, PDB code: 8co4:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Zinc where determined in the Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina, PDB code: 8co4:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Zinc binding site 1 out of 8 in 8co4
Go back to
Zinc binding site 1 out
of 8 in the Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
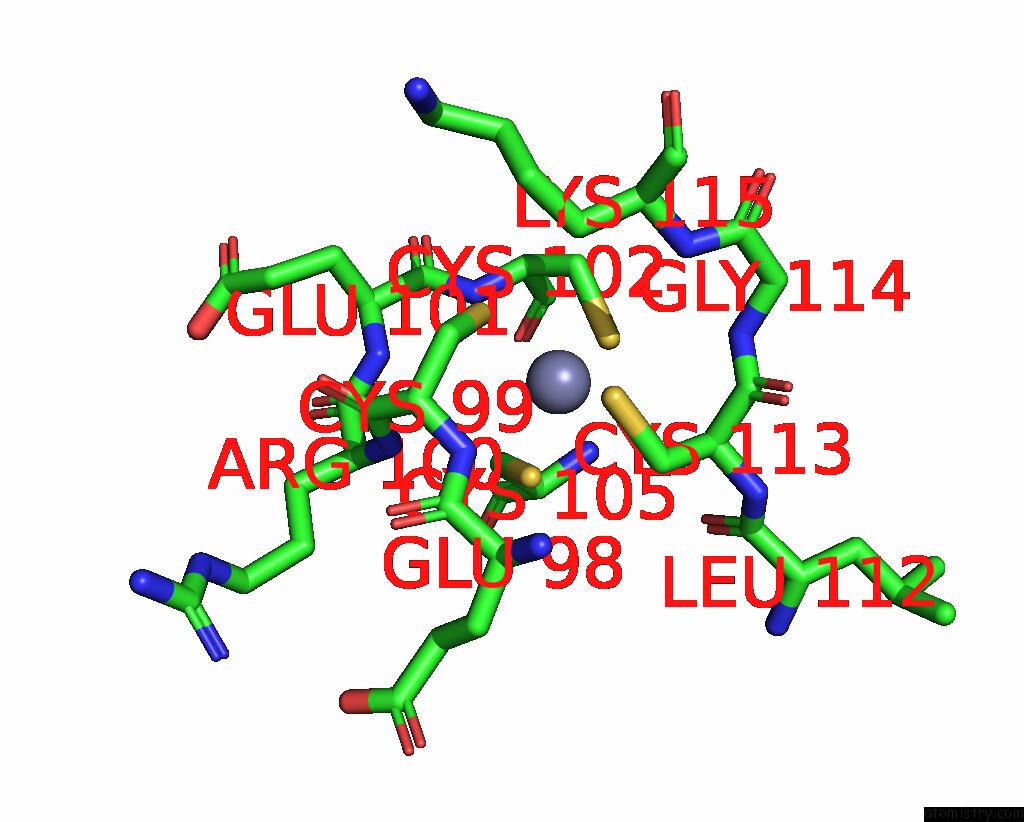
Mono view
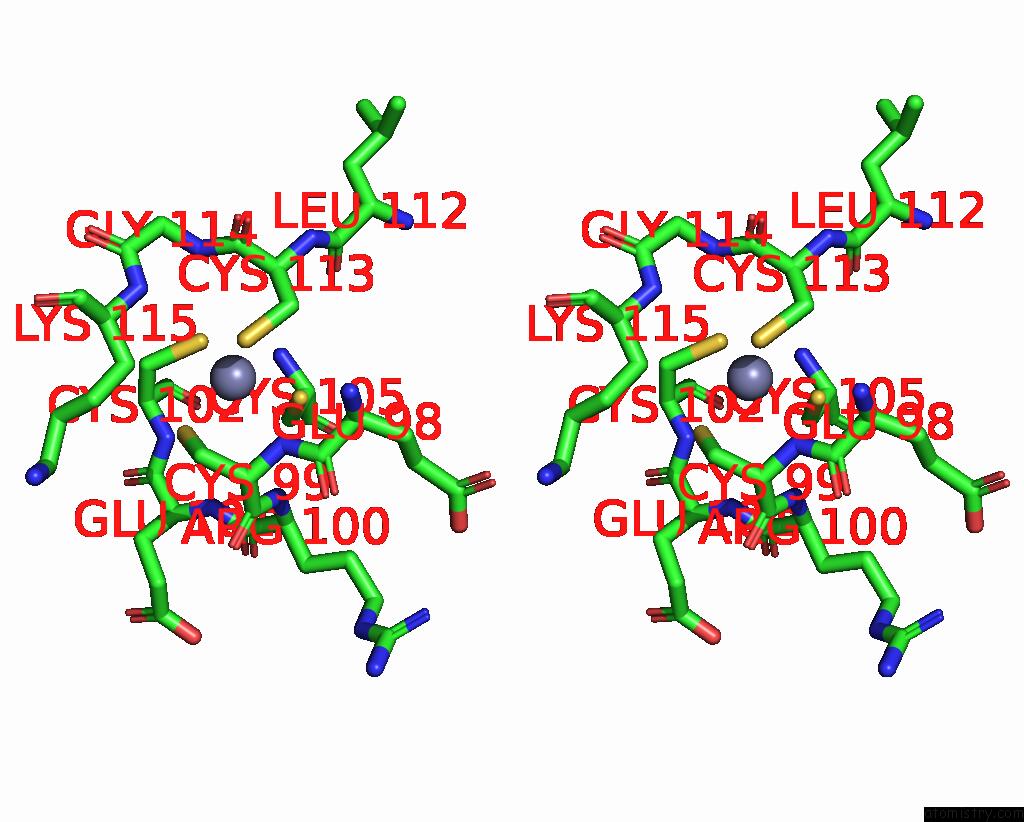
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina within 5.0Å range:
|
Zinc binding site 2 out of 8 in 8co4
Go back to
Zinc binding site 2 out
of 8 in the Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
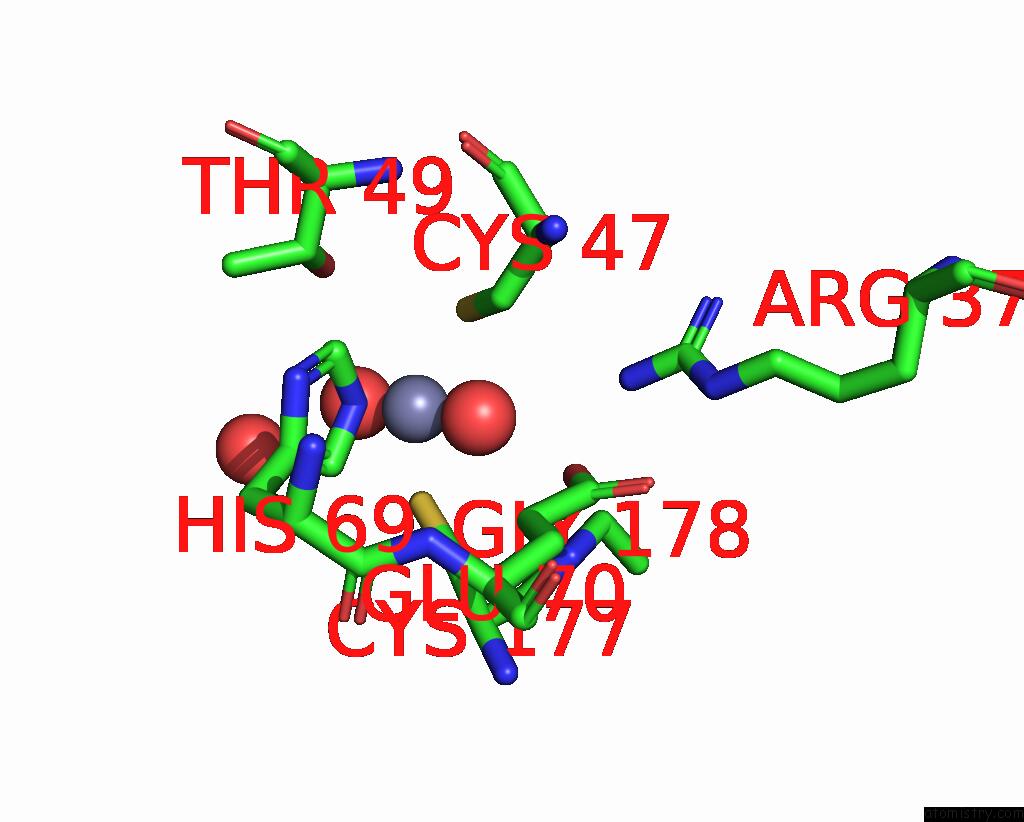
Mono view
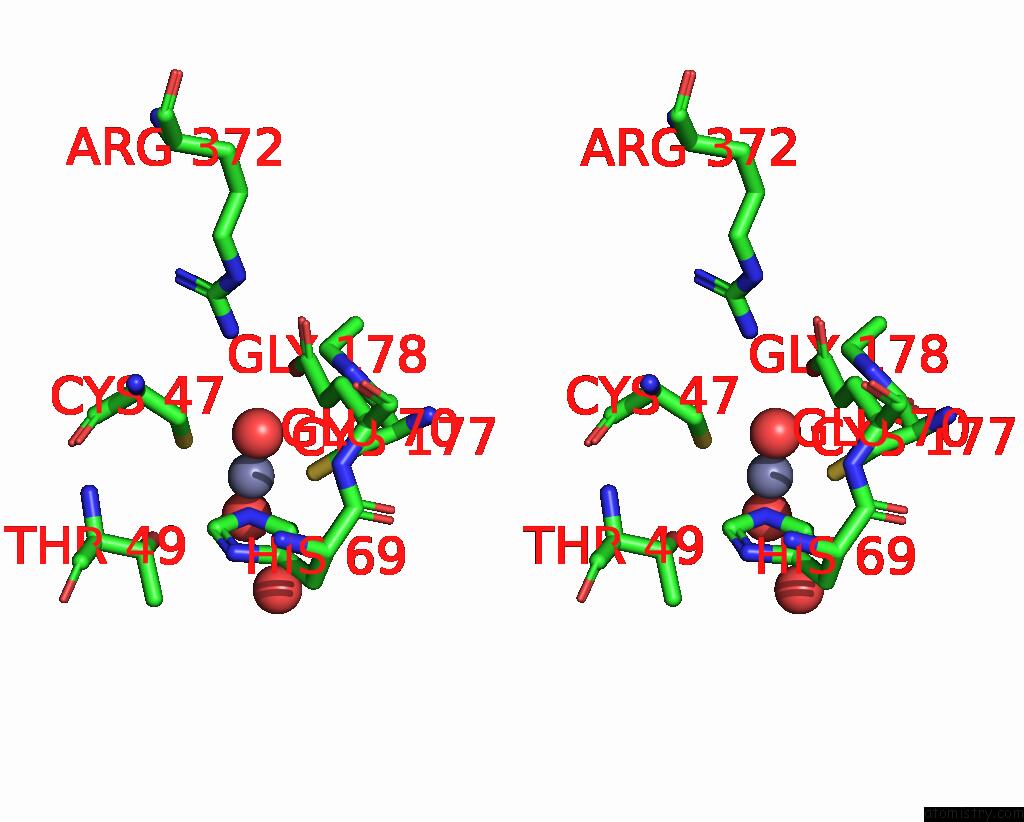
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina within 5.0Å range:
|
Zinc binding site 3 out of 8 in 8co4
Go back to
Zinc binding site 3 out
of 8 in the Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
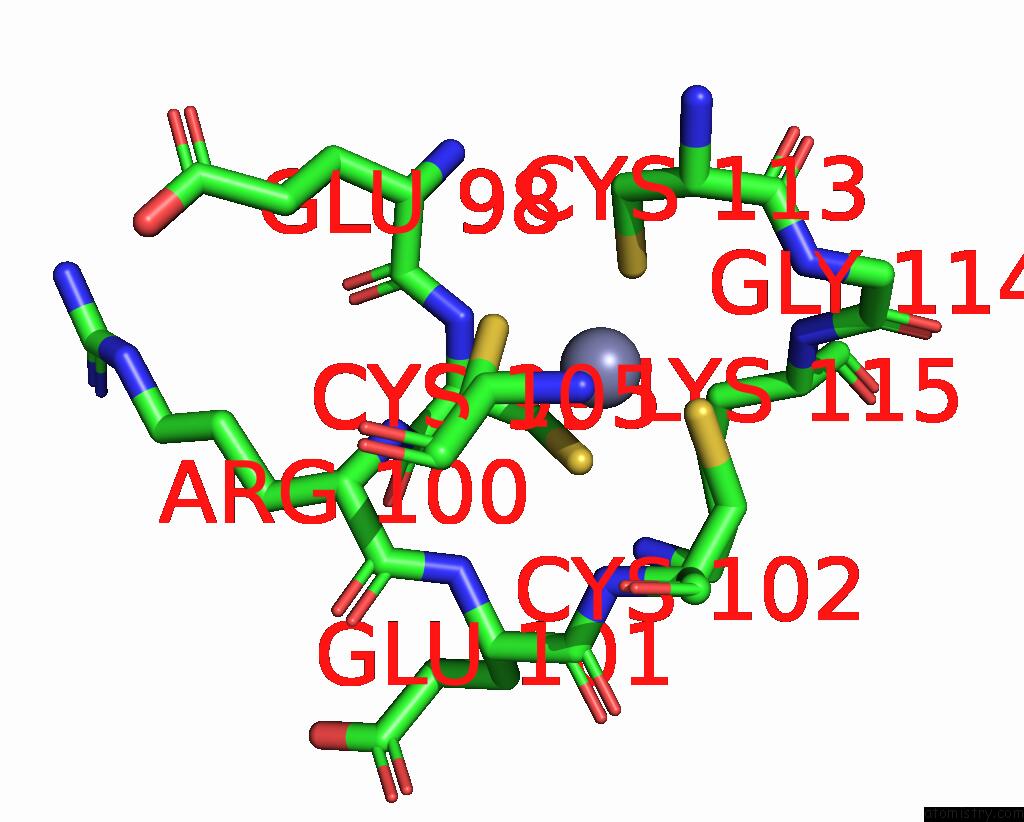
Mono view
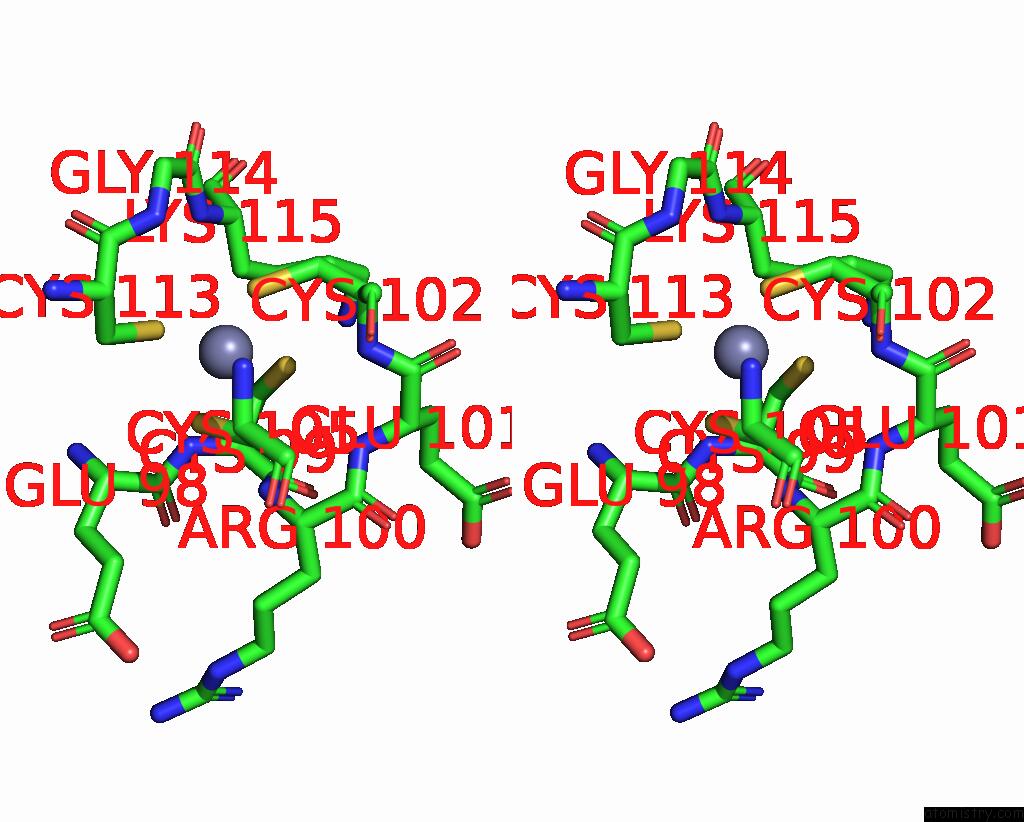
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina within 5.0Å range:
|
Zinc binding site 4 out of 8 in 8co4
Go back to
Zinc binding site 4 out
of 8 in the Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
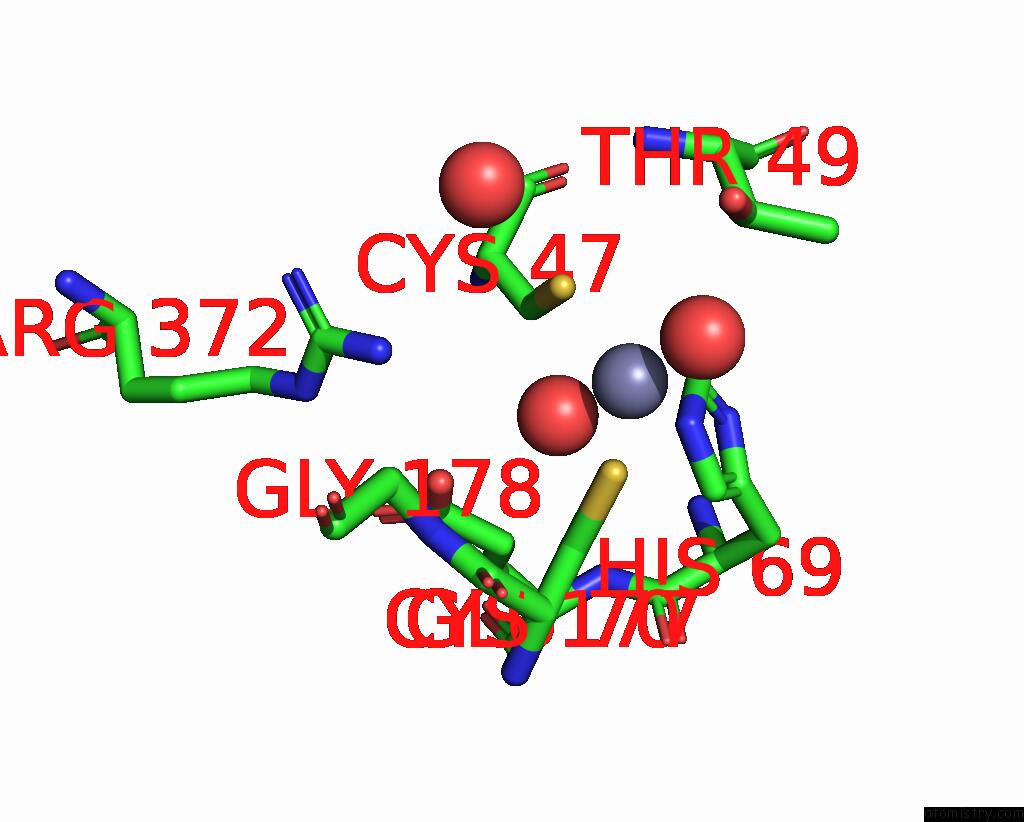
Mono view
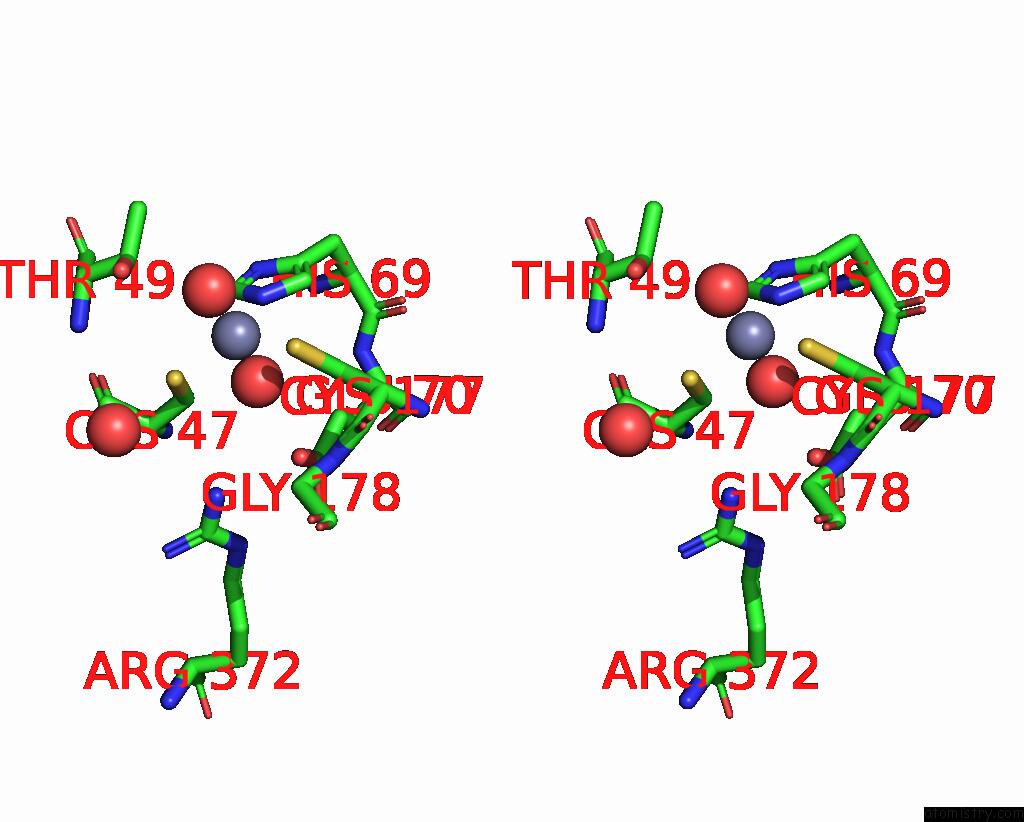
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina within 5.0Å range:
|
Zinc binding site 5 out of 8 in 8co4
Go back to
Zinc binding site 5 out
of 8 in the Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
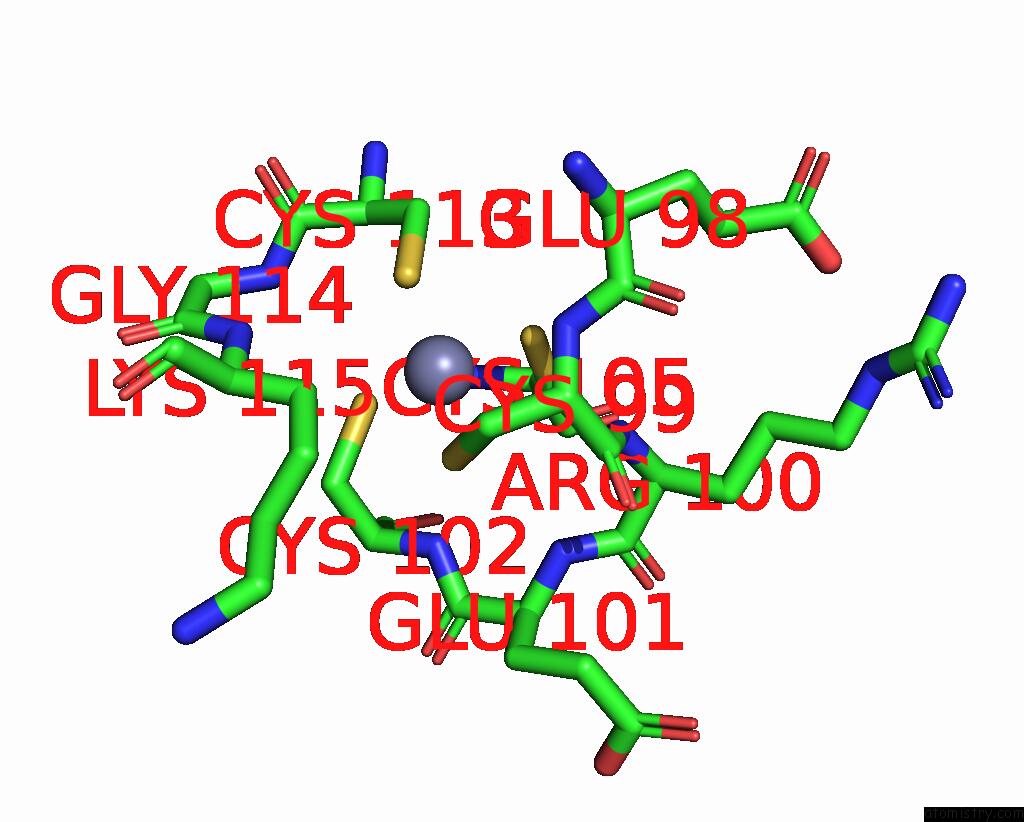
Mono view
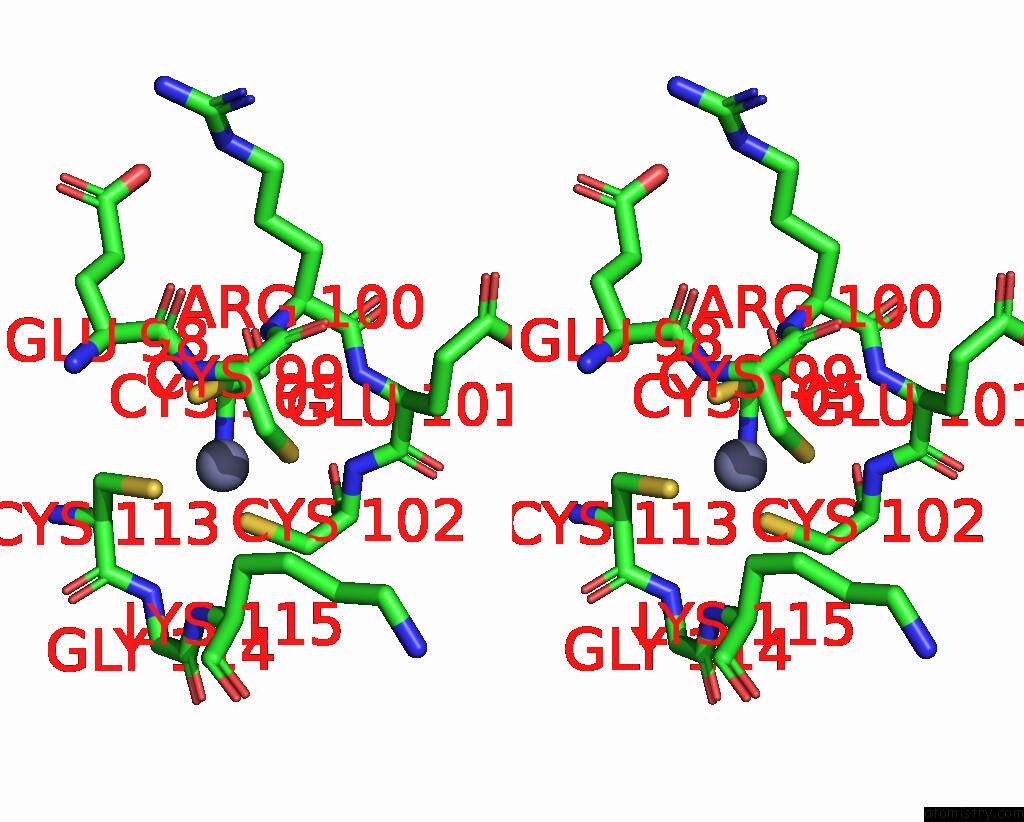
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina within 5.0Å range:
|
Zinc binding site 6 out of 8 in 8co4
Go back to
Zinc binding site 6 out
of 8 in the Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
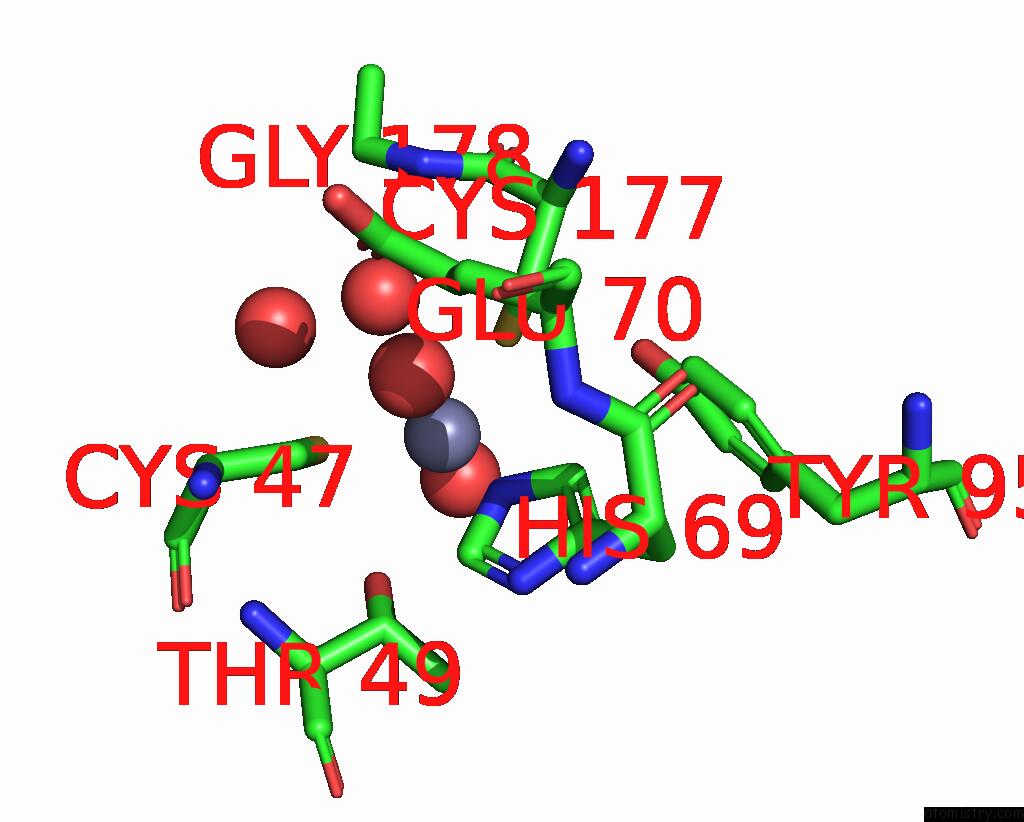
Mono view
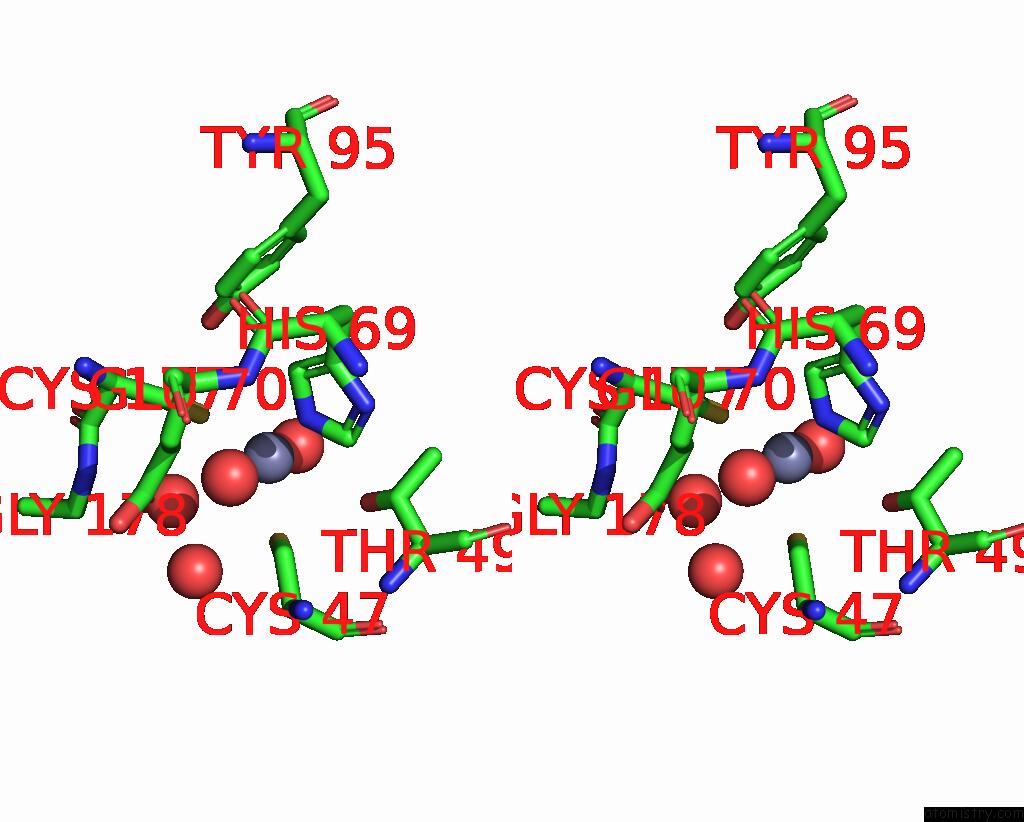
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina within 5.0Å range:
|
Zinc binding site 7 out of 8 in 8co4
Go back to
Zinc binding site 7 out
of 8 in the Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
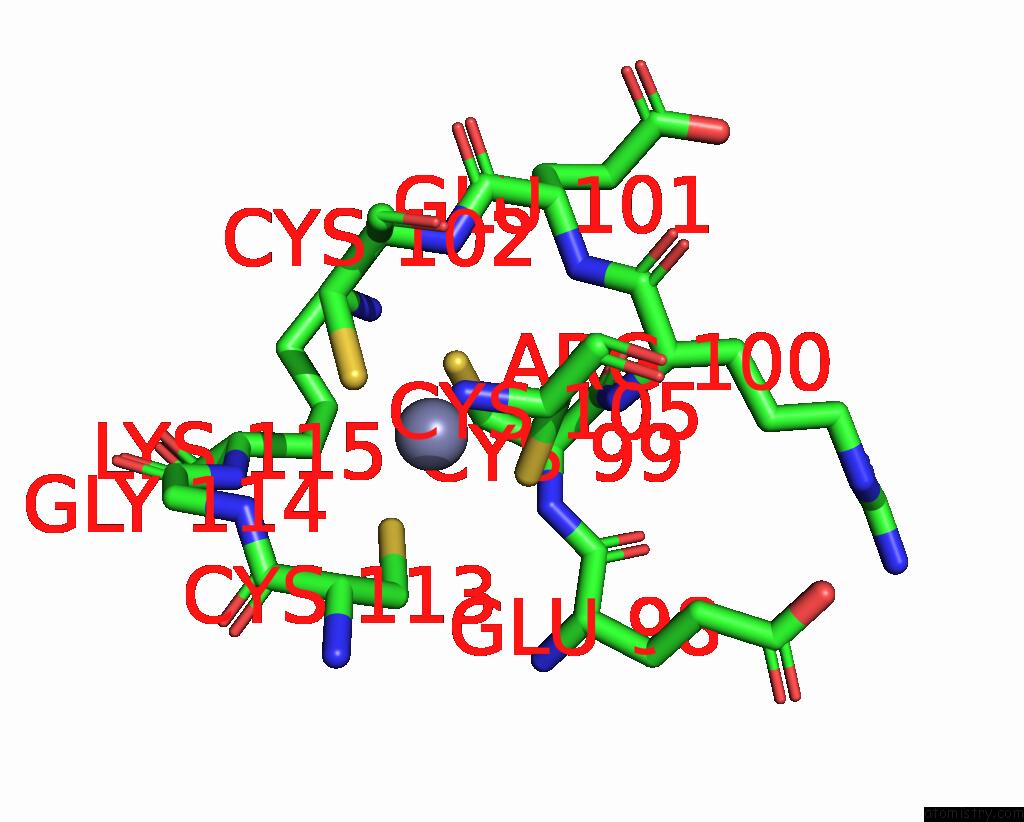
Mono view
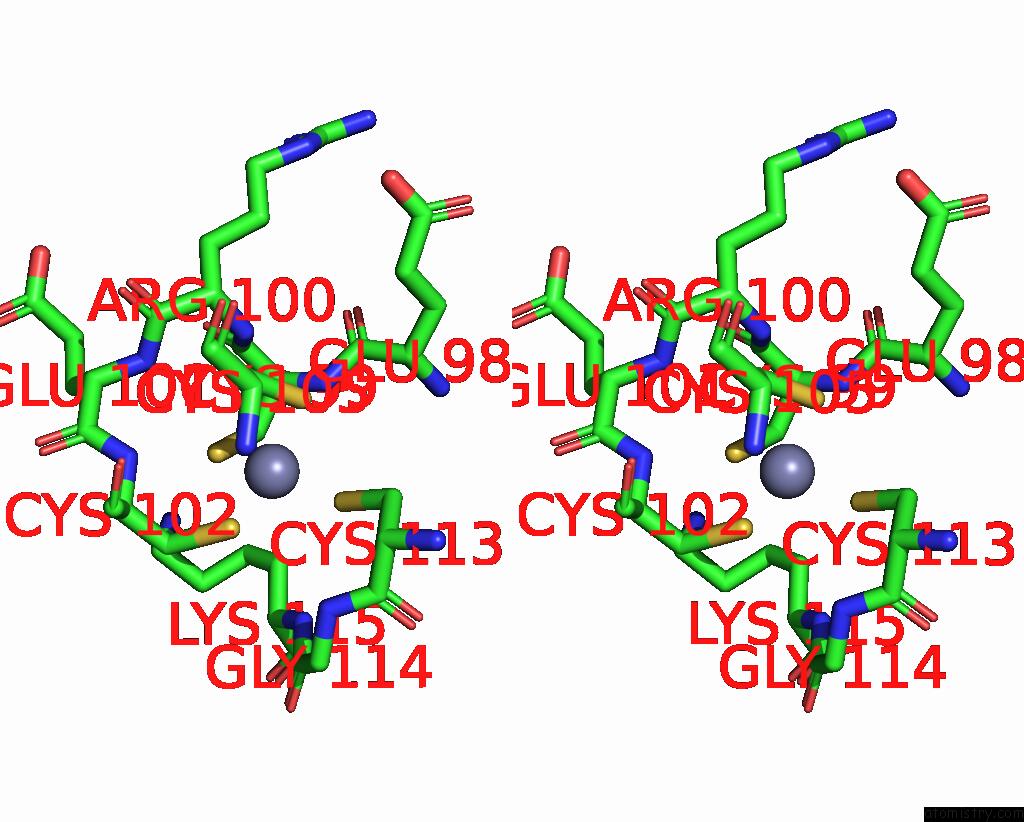
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina within 5.0Å range:
|
Zinc binding site 8 out of 8 in 8co4
Go back to
Zinc binding site 8 out
of 8 in the Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina
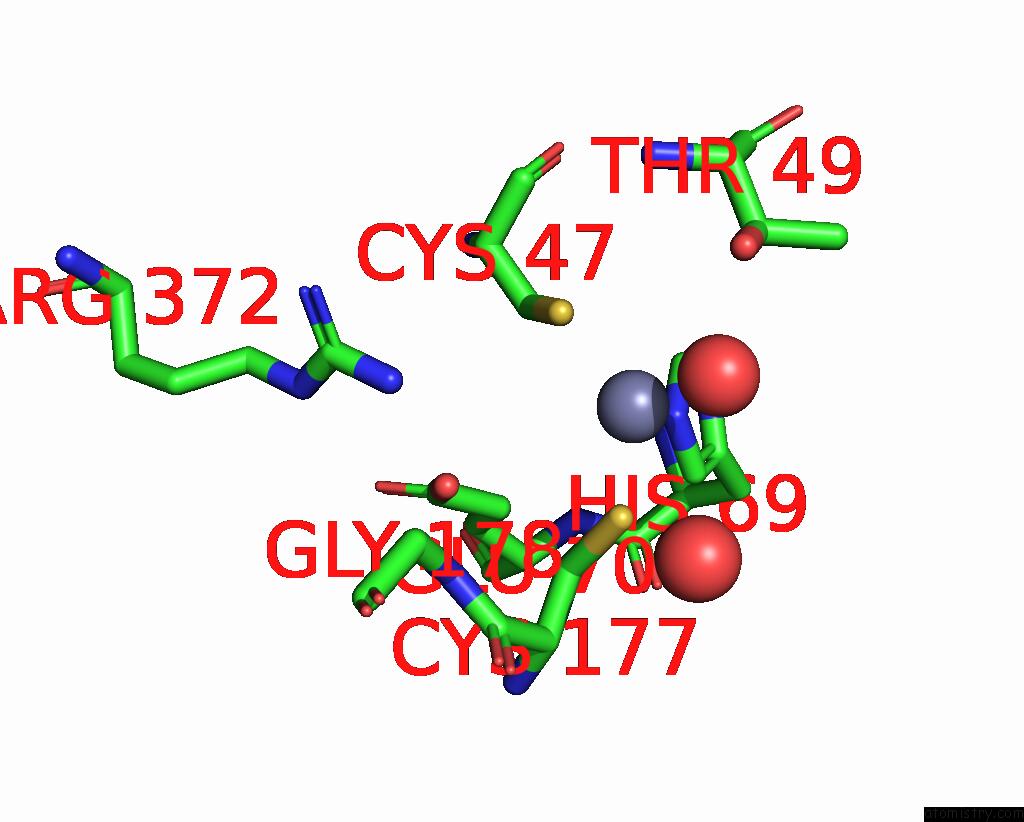
Mono view
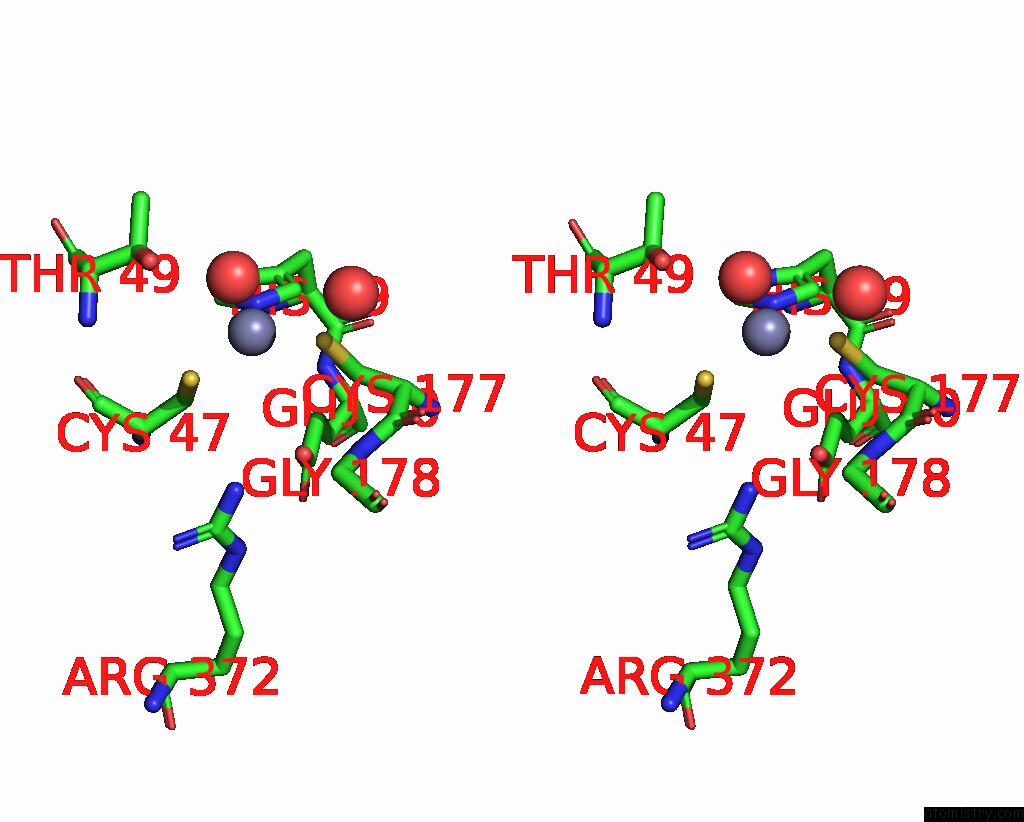
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 8 of Crystal Structure of Apo S-Nitrosoglutathione Reductase From Arabidopsis Thalina within 5.0Å range:
|
Reference:
M.Meloni,
J.Rossi,
S.Fanti,
G.Carloni,
D.Tedesco,
P.Treffon,
L.Piccinini,
G.Falini,
P.Trost,
E.Vierling,
F.Licausi,
B.Giuntoli,
F.Musiani,
S.Fermani,
M.Zaffagnini.
Structural and Biochemical Characterization of Arabidopsis Alcohol Dehydrogenases Reveals Distinct Functional Properties But Similar Redox Sensitivity. Plant J. 2024.
ISSN: ESSN 1365-313X
PubMed: 38308388
DOI: 10.1111/TPJ.16651
Page generated: Fri Aug 22 09:01:06 2025
ISSN: ESSN 1365-313X
PubMed: 38308388
DOI: 10.1111/TPJ.16651
Last articles
Zn in 9CDHZn in 9CCH
Zn in 9CDG
Zn in 9CDF
Zn in 9CBT
Zn in 9C8U
Zn in 9CBR
Zn in 9CBP
Zn in 9CAH
Zn in 9CBQ