Zinc »
PDB 8cdb-8cro »
8cli »
Zinc in PDB 8cli: Tfiiic Taub-Dna Monomer
Enzymatic activity of Tfiiic Taub-Dna Monomer
All present enzymatic activity of Tfiiic Taub-Dna Monomer:
2.3.1.48;
2.3.1.48;
Zinc Binding Sites:
The binding sites of Zinc atom in the Tfiiic Taub-Dna Monomer
(pdb code 8cli). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Tfiiic Taub-Dna Monomer, PDB code: 8cli:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Tfiiic Taub-Dna Monomer, PDB code: 8cli:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 8cli
Go back to
Zinc binding site 1 out
of 2 in the Tfiiic Taub-Dna Monomer
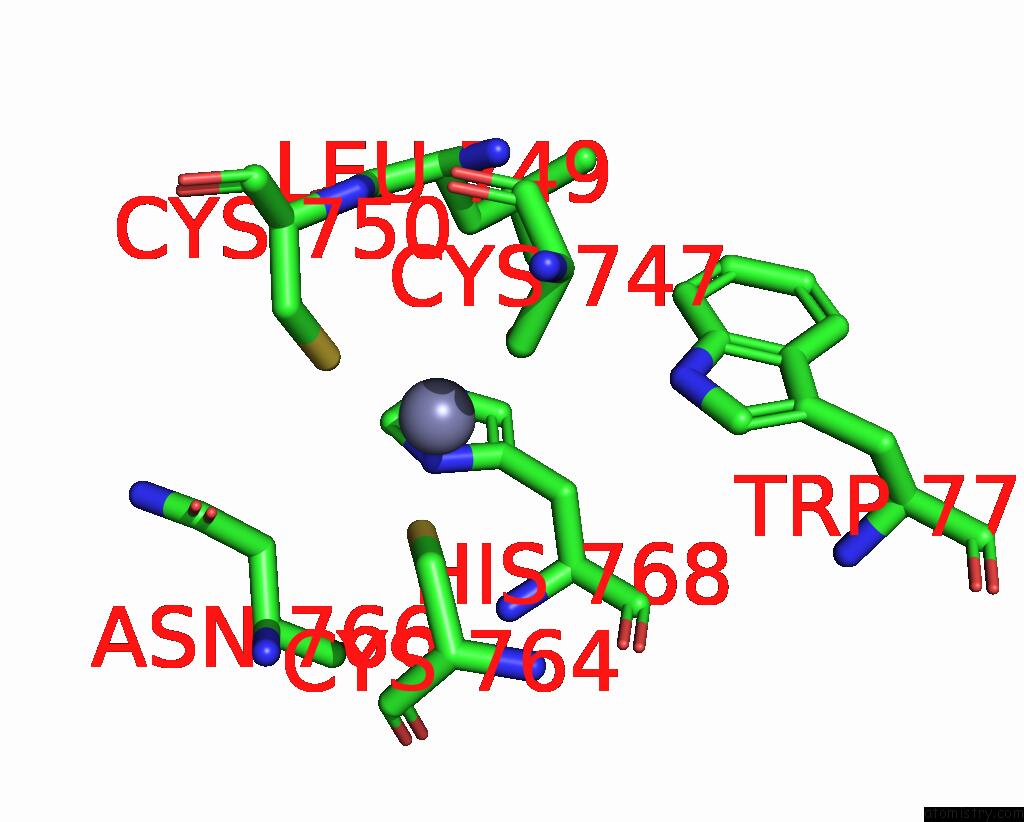
Mono view
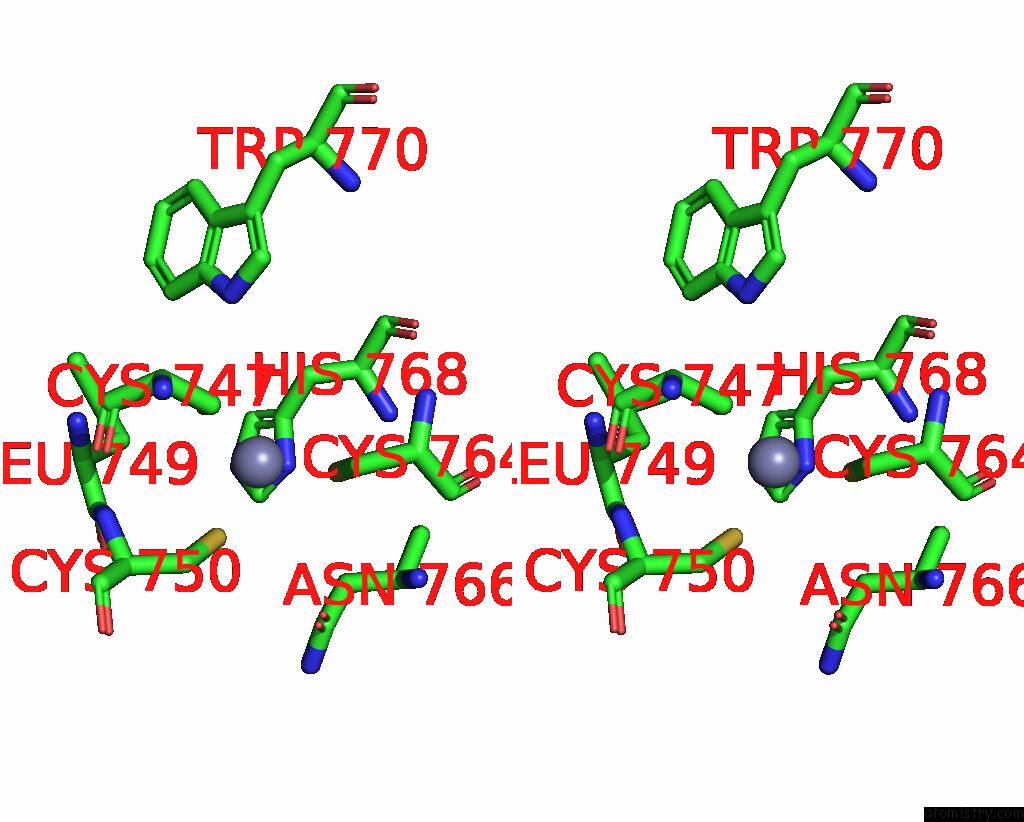
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Tfiiic Taub-Dna Monomer within 5.0Å range:
|
Zinc binding site 2 out of 2 in 8cli
Go back to
Zinc binding site 2 out
of 2 in the Tfiiic Taub-Dna Monomer
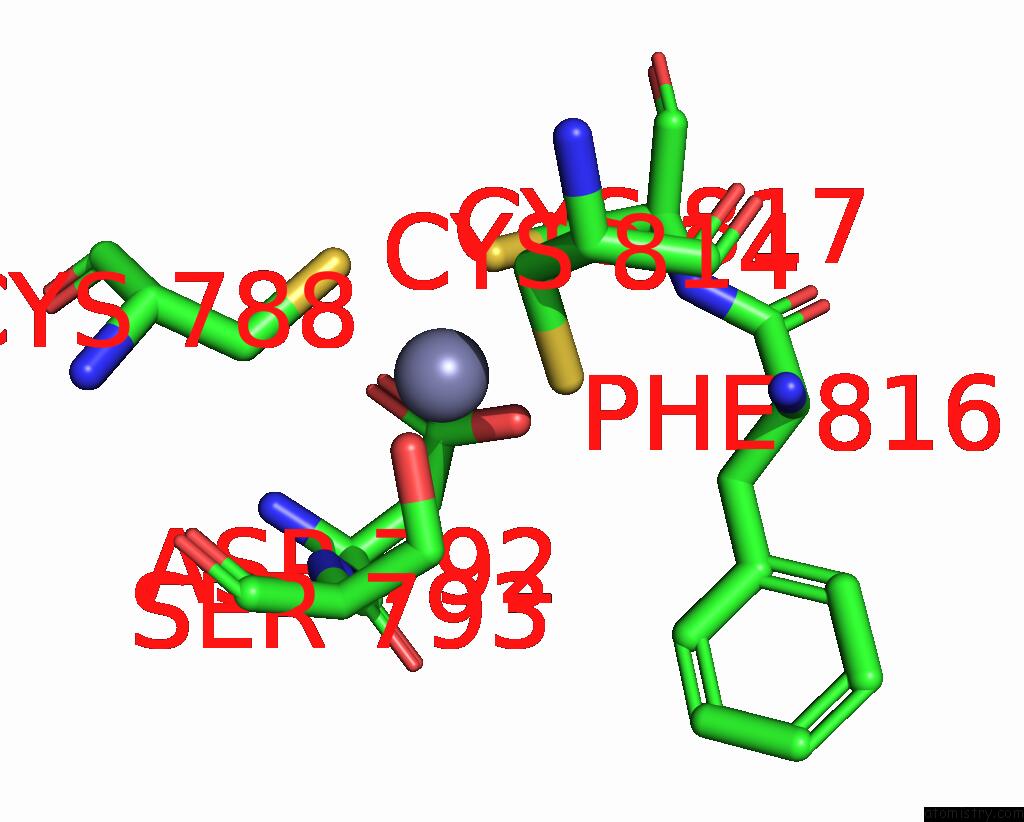
Mono view
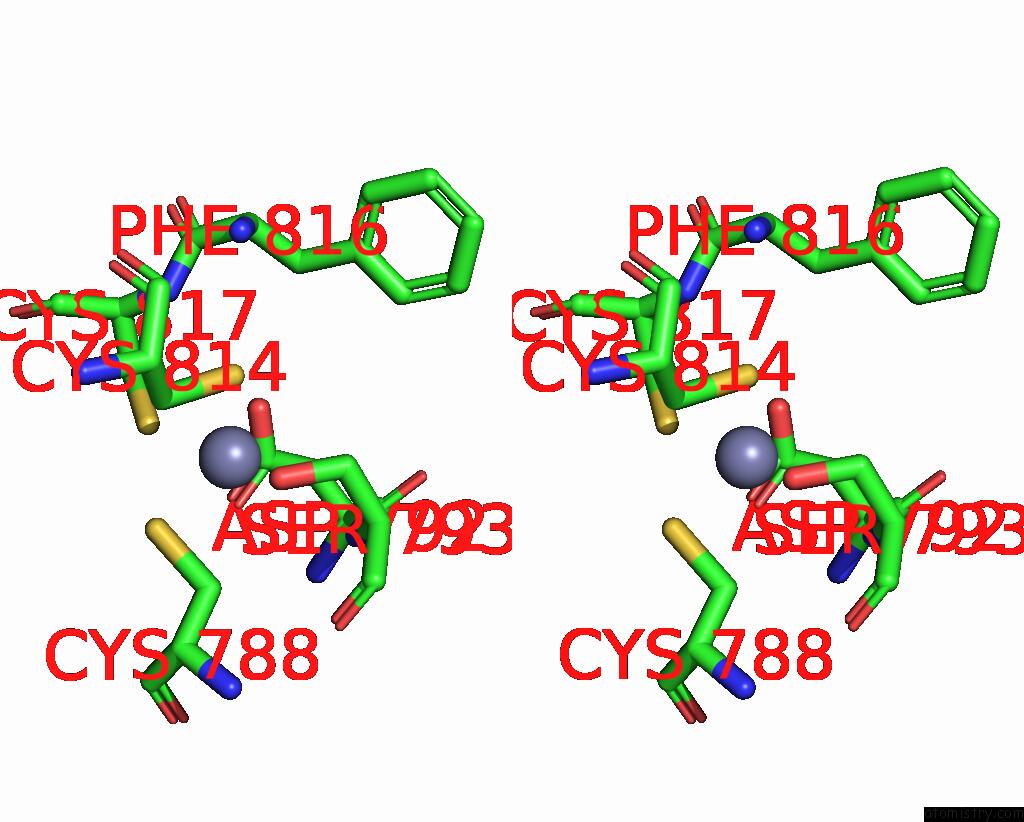
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Tfiiic Taub-Dna Monomer within 5.0Å range:
|
Reference:
S.D.Wolfram,
G.Mathias,
H.Luis,
H.Thomas,
E.Sebastian,
M.Christoph.
Cryo-Em Structures of Human Tfiiic Reveal A Multi-Step Mechanism For Trna Gene Promoter Recognition To Be Published.
Page generated: Fri Aug 22 08:57:43 2025
Last articles
Zn in 8QUEZn in 8QUC
Zn in 8QVO
Zn in 8QU8
Zn in 8QU5
Zn in 8QU6
Zn in 8QTK
Zn in 8QTU
Zn in 8QTI
Zn in 8QTH