Zinc »
PDB 7ygi-7yrs »
7ygw »
Zinc in PDB 7ygw: Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2
Protein crystallography data
The structure of Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2, PDB code: 7ygw
was solved by
S.A.Mun,
J.Park,
J.Y.Kang,
T.Park,
M.Jin,
J.Yang,
S.H.Eom,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.96 / 1.72 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 43.959, 47.393, 63.399, 90, 90, 90 |
| R / Rfree (%) | 19.8 / 21.1 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2
(pdb code 7ygw). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2, PDB code: 7ygw:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2, PDB code: 7ygw:
Jump to Zinc binding site number: 1; 2; 3; 4;
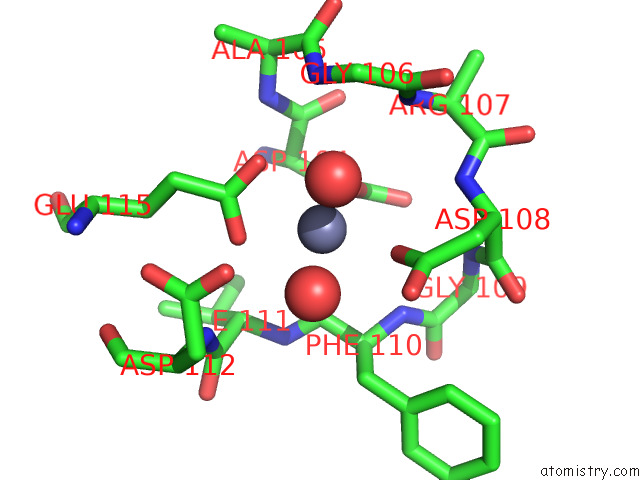
Zinc binding site 1 out of 4 in 7ygw
Go back to
Zinc binding site 1 out
of 4 in the Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2 within 5.0Å range:
|
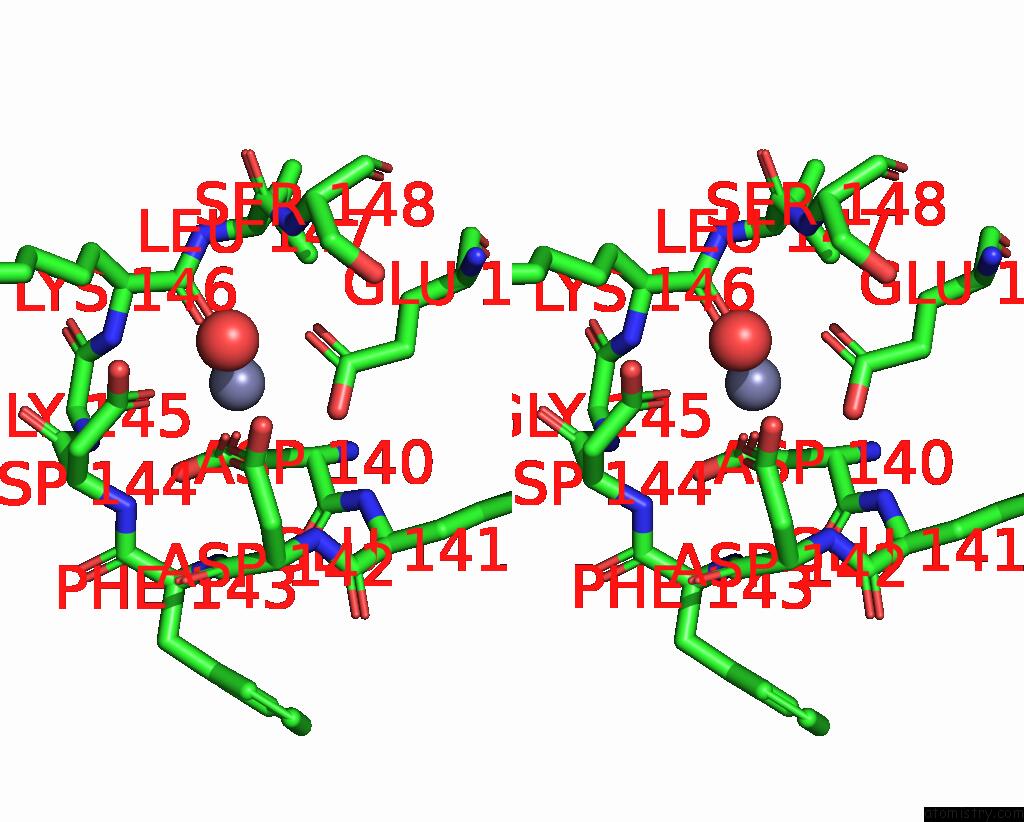
Zinc binding site 2 out of 4 in 7ygw
Go back to
Zinc binding site 2 out
of 4 in the Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2 within 5.0Å range:
|
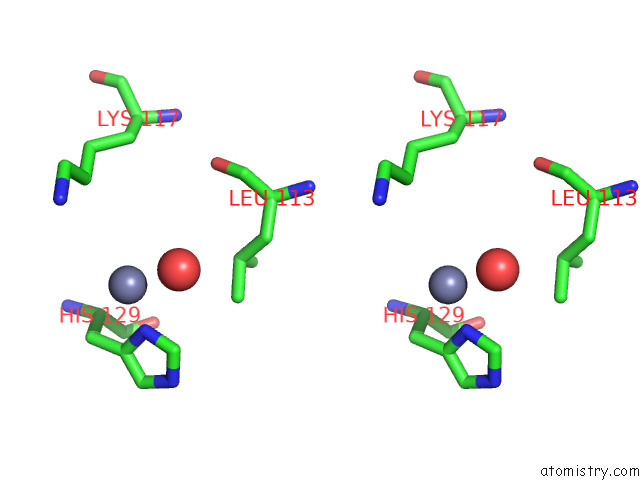
Zinc binding site 3 out of 4 in 7ygw
Go back to
Zinc binding site 3 out
of 4 in the Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2 within 5.0Å range:
|
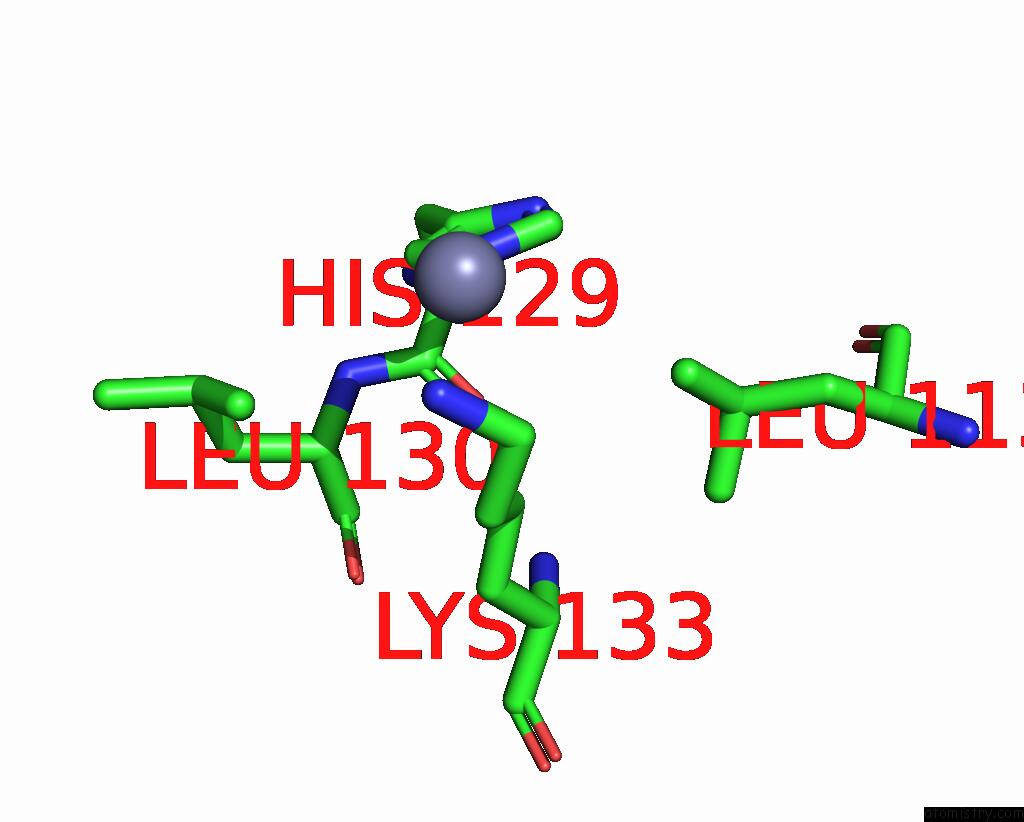
Zinc binding site 4 out of 4 in 7ygw
Go back to
Zinc binding site 4 out
of 4 in the Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2
Mono view
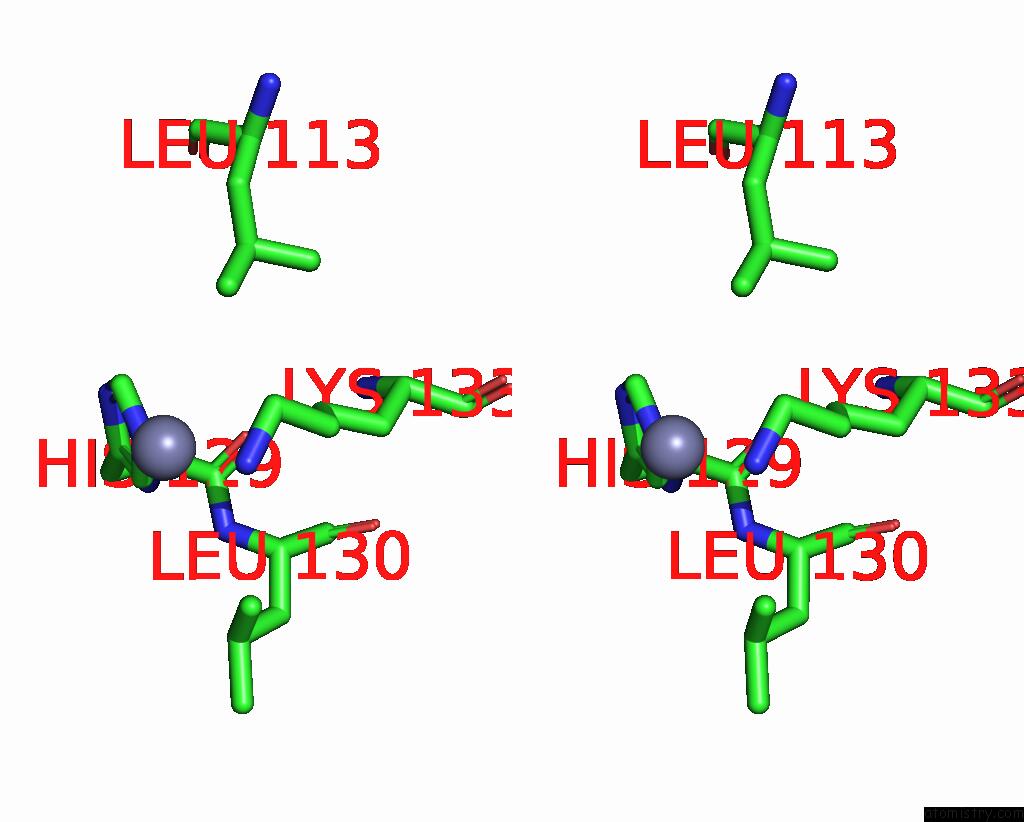
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of the ZN2+-Bound EFHD1/Swiprosin-2 within 5.0Å range:
|
Reference:
S.A.Mun,
J.Park,
J.Y.Kang,
T.Park,
M.Jin,
J.Yang,
S.H.Eom.
Structural and Biochemical Insights Into Zn 2+ -Bound Ef-Hand Proteins, EFHD1 and EFHD2. Iucrj V. 10 233 2023.
ISSN: ESSN 2052-2525
PubMed: 36862489
DOI: 10.1107/S2052252523001501
Page generated: Fri Aug 22 07:05:03 2025
ISSN: ESSN 2052-2525
PubMed: 36862489
DOI: 10.1107/S2052252523001501
Last articles
Zn in 8J62Zn in 8J4B
Zn in 8J56
Zn in 8J2P
Zn in 8J49
Zn in 8J48
Zn in 8J25
Zn in 8J3R
Zn in 8J2O
Zn in 8IUH