Zinc »
PDB 7t85-7ts7 »
7to1 »
Zinc in PDB 7to1: Cryo-Em Structure of Rig-I Bound to the End of P3SLR30 (+Atp)
Enzymatic activity of Cryo-Em Structure of Rig-I Bound to the End of P3SLR30 (+Atp)
All present enzymatic activity of Cryo-Em Structure of Rig-I Bound to the End of P3SLR30 (+Atp):
3.6.4.13;
3.6.4.13;
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryo-Em Structure of Rig-I Bound to the End of P3SLR30 (+Atp)
(pdb code 7to1). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Cryo-Em Structure of Rig-I Bound to the End of P3SLR30 (+Atp), PDB code: 7to1:
In total only one binding site of Zinc was determined in the Cryo-Em Structure of Rig-I Bound to the End of P3SLR30 (+Atp), PDB code: 7to1:
Zinc binding site 1 out of 1 in 7to1
Go back to
Zinc binding site 1 out
of 1 in the Cryo-Em Structure of Rig-I Bound to the End of P3SLR30 (+Atp)
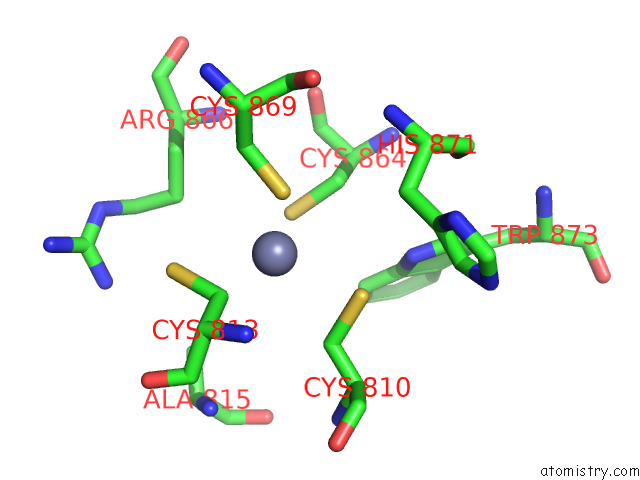
Mono view
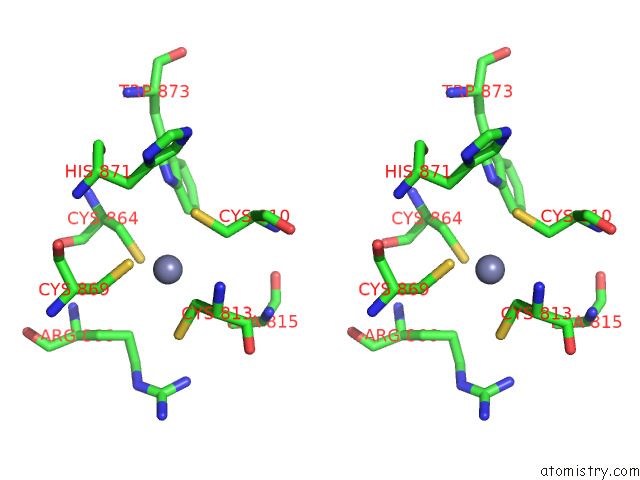
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryo-Em Structure of Rig-I Bound to the End of P3SLR30 (+Atp) within 5.0Å range:
|
Reference:
W.Wang,
A.M.Pyle.
The Rig-I Receptor Adopts Two Different Conformations For Distinguishing Host From Viral Rna Ligands. Mol.Cell V. 82 4131 2022.
ISSN: ISSN 1097-2765
PubMed: 36272408
DOI: 10.1016/J.MOLCEL.2022.09.029
Page generated: Fri Aug 22 05:01:47 2025
ISSN: ISSN 1097-2765
PubMed: 36272408
DOI: 10.1016/J.MOLCEL.2022.09.029
Last articles
Zn in 8DPRZn in 8DPO
Zn in 8DPC
Zn in 8DJH
Zn in 8DJI
Zn in 8DI4
Zn in 8DJ9
Zn in 8DEY
Zn in 8DGT
Zn in 8DH6