Zinc »
PDB 7t85-7ts7 »
7t92 »
Zinc in PDB 7t92: Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex
(pdb code 7t92). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 5 binding sites of Zinc where determined in the Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex, PDB code: 7t92:
Jump to Zinc binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Zinc where determined in the Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex, PDB code: 7t92:
Jump to Zinc binding site number: 1; 2; 3; 4; 5;
Zinc binding site 1 out of 5 in 7t92
Go back to
Zinc binding site 1 out
of 5 in the Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex
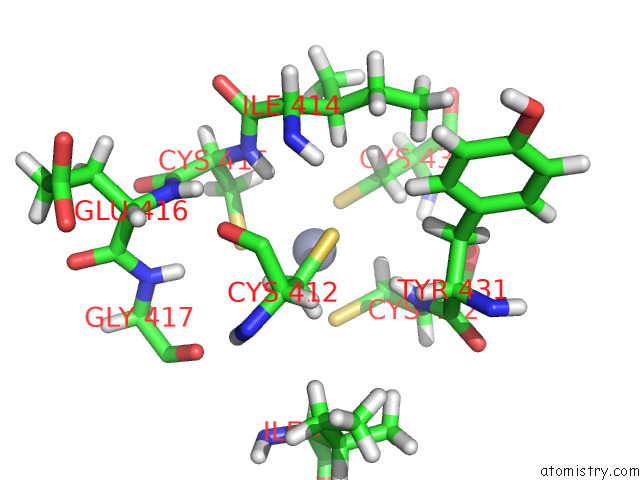
Mono view
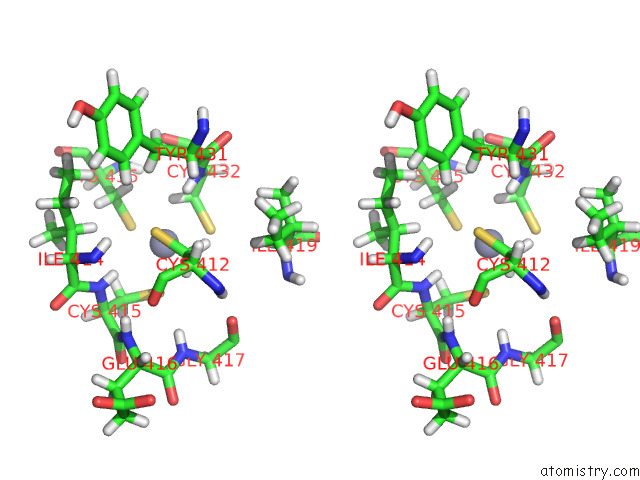
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex within 5.0Å range:
|
Zinc binding site 2 out of 5 in 7t92
Go back to
Zinc binding site 2 out
of 5 in the Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex
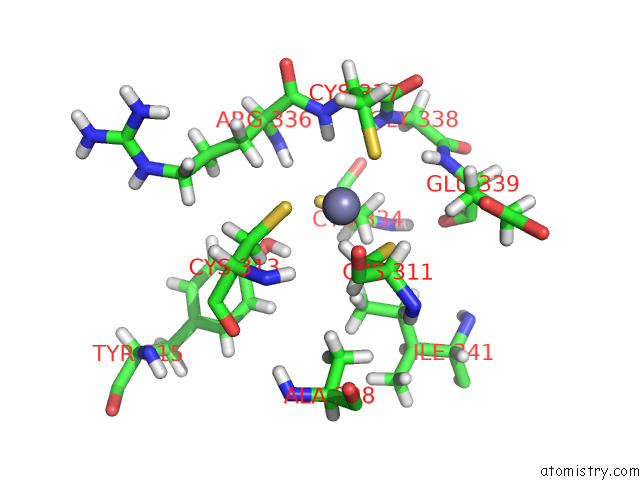
Mono view
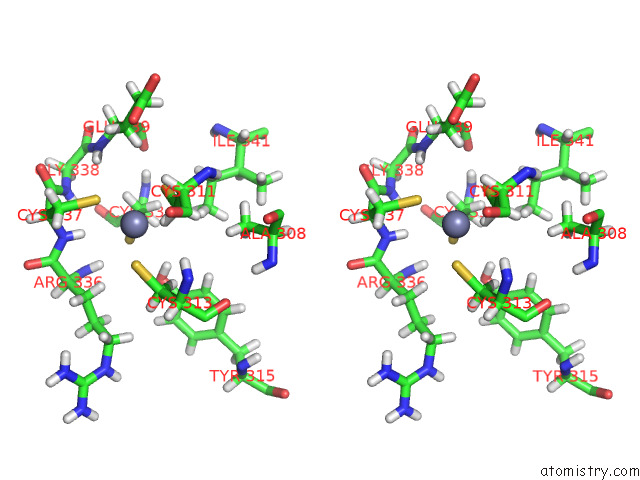
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex within 5.0Å range:
|
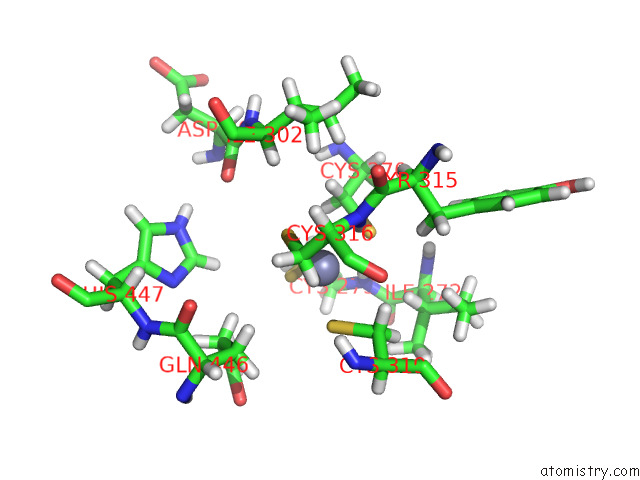
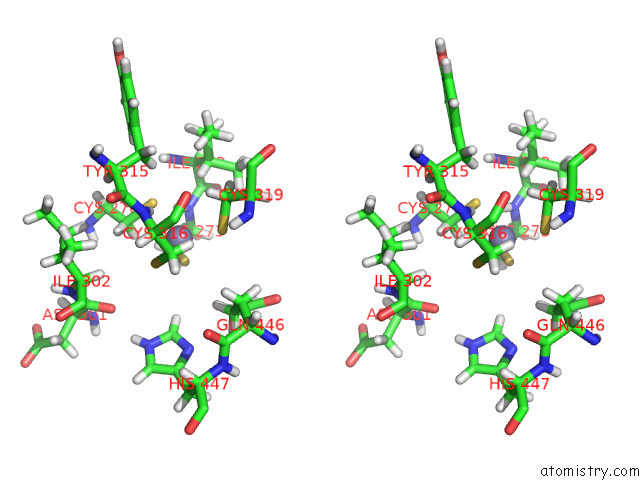
Zinc binding site 3 out of 5 in 7t92
Go back to
Zinc binding site 3 out
of 5 in the Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex within 5.0Å range:
|
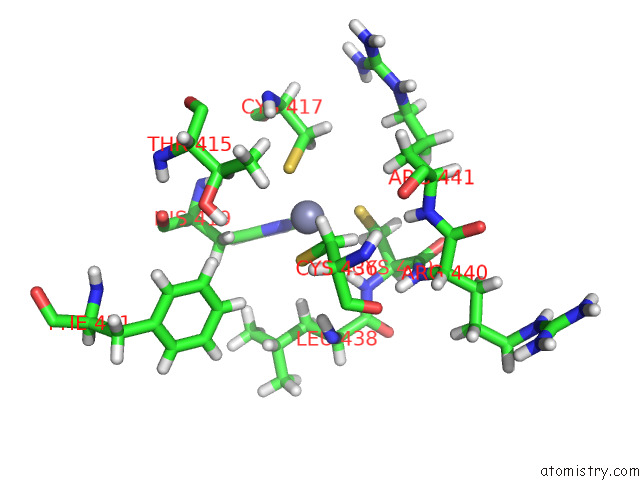
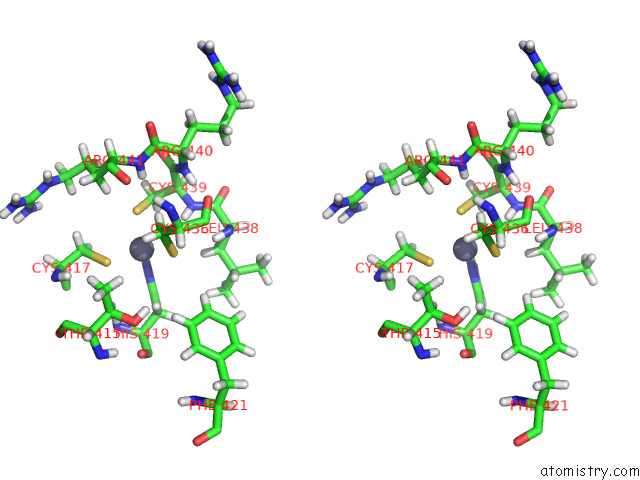
Zinc binding site 4 out of 5 in 7t92
Go back to
Zinc binding site 4 out
of 5 in the Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex within 5.0Å range:
|
Zinc binding site 5 out of 5 in 7t92
Go back to
Zinc binding site 5 out
of 5 in the Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex
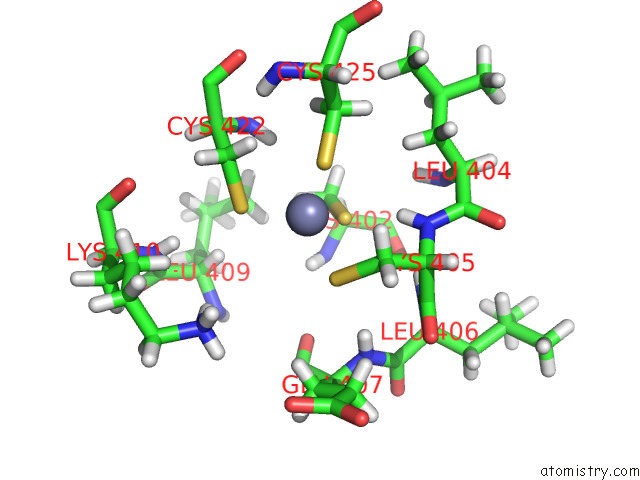
Mono view
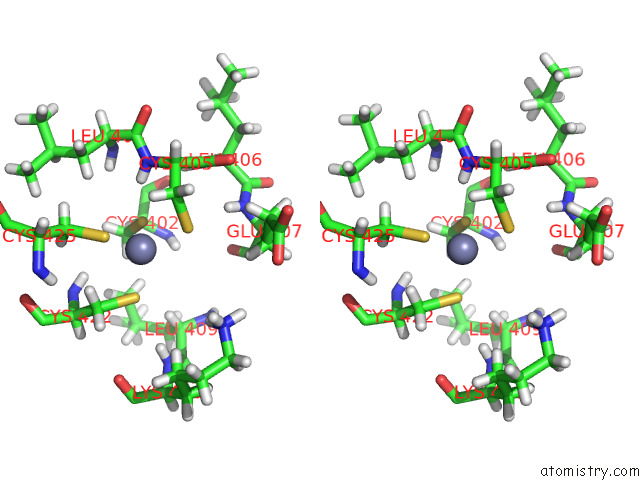
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Structure of the Peroxisomal Retro-Translocon Formed By A Heterotrimeric Ubiquitin Ligase Complex within 5.0Å range:
|
Reference:
P.Feng,
X.Wu,
S.K.Erramilli,
J.A.Paulo,
P.Knejski,
S.P.Gygi,
A.A.Kossiakoff,
T.A.Rapoport.
A Peroxisomal Ubiquitin Ligase Complex Forms A Retrotranslocation Channel. Nature V. 607 374 2022.
ISSN: ESSN 1476-4687
PubMed: 35768507
DOI: 10.1038/S41586-022-04903-X
Page generated: Fri Aug 22 04:56:23 2025
ISSN: ESSN 1476-4687
PubMed: 35768507
DOI: 10.1038/S41586-022-04903-X
Last articles
Zn in 8DZFZn in 8DZE
Zn in 8DZJ
Zn in 8DY9
Zn in 8DY7
Zn in 8DYQ
Zn in 8DWL
Zn in 8DWJ
Zn in 8DQN
Zn in 8DW6