Zinc »
PDB 7rbr-7rre »
7rm6 »
Zinc in PDB 7rm6: Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide
Enzymatic activity of Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide
All present enzymatic activity of Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide:
1.1.1.1;
1.1.1.1;
Protein crystallography data
The structure of Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide, PDB code: 7rm6
was solved by
C.Zheng,
S.G.Boxer,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.94 / 1.43 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 44.142, 50.979, 92.864, 92.41, 103.03, 108.94 |
| R / Rfree (%) | 19 / 20.8 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide
(pdb code 7rm6). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide, PDB code: 7rm6:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide, PDB code: 7rm6:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 7rm6
Go back to
Zinc binding site 1 out
of 4 in the Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide
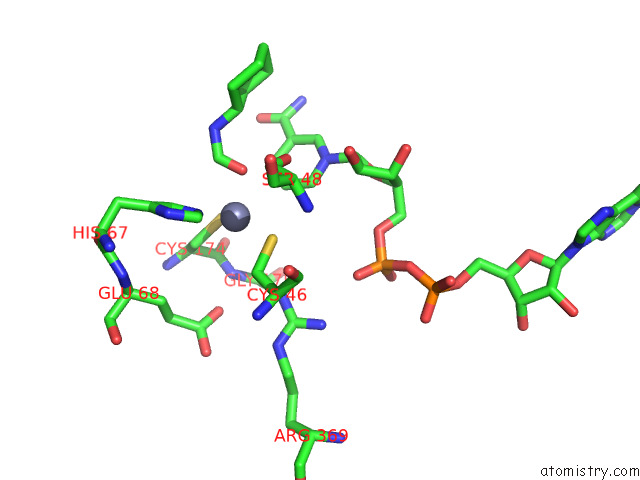
Mono view
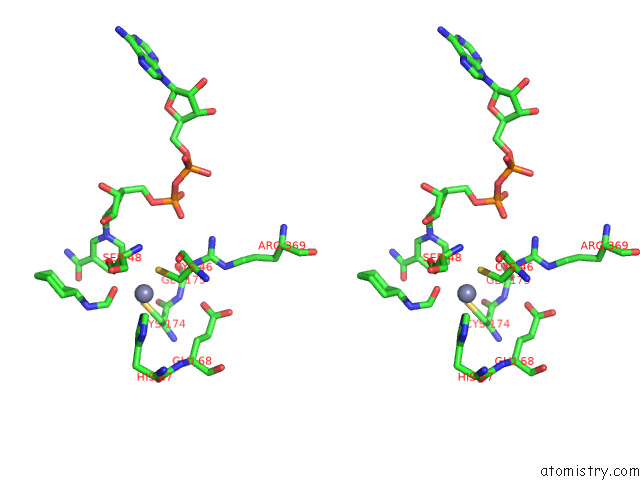
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide within 5.0Å range:
|
Zinc binding site 2 out of 4 in 7rm6
Go back to
Zinc binding site 2 out
of 4 in the Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide
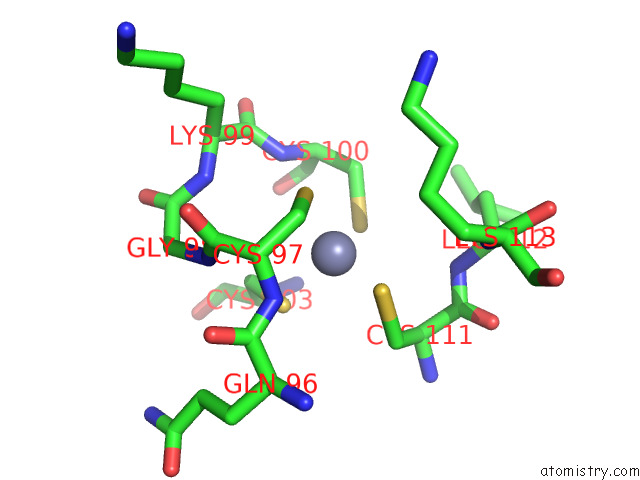
Mono view
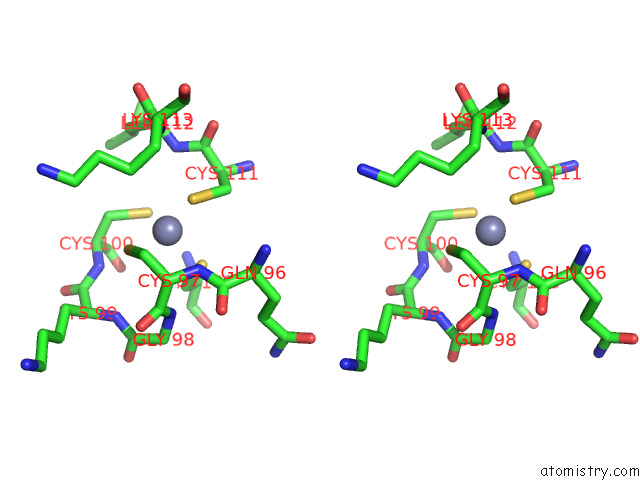
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide within 5.0Å range:
|
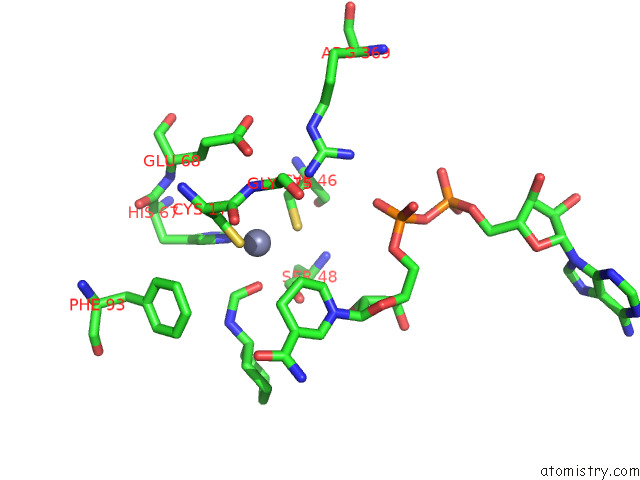
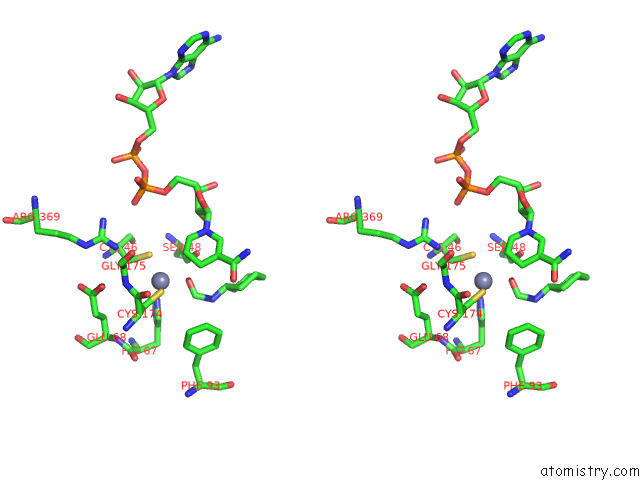
Zinc binding site 3 out of 4 in 7rm6
Go back to
Zinc binding site 3 out
of 4 in the Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide within 5.0Å range:
|
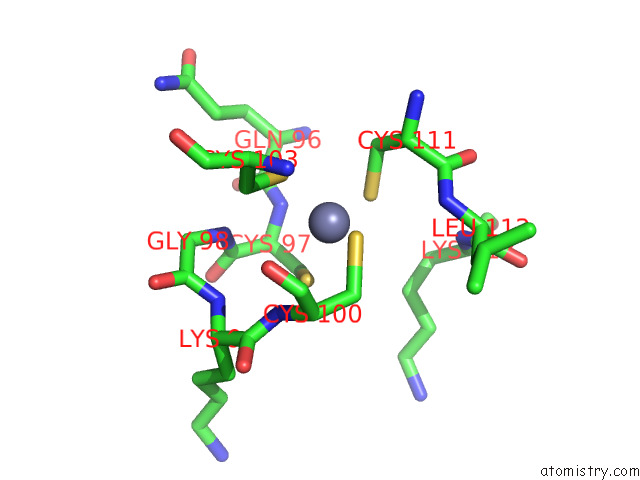
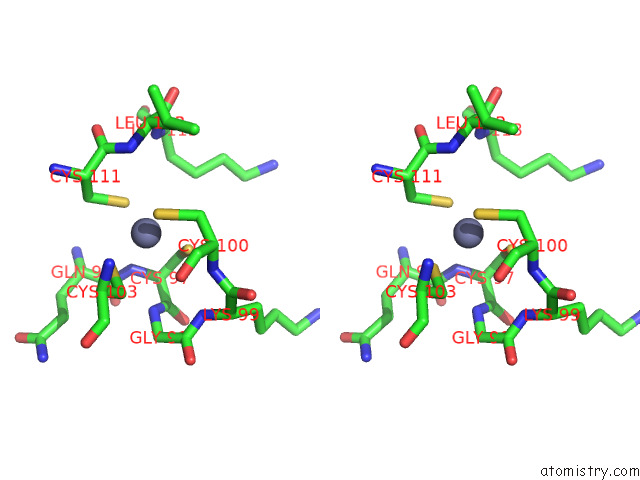
Zinc binding site 4 out of 4 in 7rm6
Go back to
Zinc binding site 4 out
of 4 in the Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Horse Liver Alcohol Dehydrogenase in Complex with Nadh and N- Cylcohexyl Formamide within 5.0Å range:
|
Reference:
C.Zheng,
Y.Mao,
J.Kozuch,
A.O.Atsango,
Z.Ji,
T.E.Markland,
S.G.Boxer.
A Two-Directional Vibrational Probe Reveals Different Electric Field Orientations in Solution and An Enzyme Active Site. Nat.Chem. V. 14 891 2022.
ISSN: ESSN 1755-4349
PubMed: 35513508
DOI: 10.1038/S41557-022-00937-W
Page generated: Fri Aug 22 04:18:02 2025
ISSN: ESSN 1755-4349
PubMed: 35513508
DOI: 10.1038/S41557-022-00937-W
Last articles
Zn in 8CLJZn in 8CLL
Zn in 8CLI
Zn in 8CJ7
Zn in 8CHX
Zn in 8CDB
Zn in 8CEF
Zn in 8CGW
Zn in 8CGV
Zn in 8CGP