Zinc »
PDB 7pcp-7pp2 »
7poo »
Zinc in PDB 7poo: Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121.
Protein crystallography data
The structure of Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121., PDB code: 7poo
was solved by
U.Eckhard,
T.Guevara,
F.X.Gomis-Ruth,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 73.47 / 2.70 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 69.55, 83.06, 157.48, 90, 90, 90 |
| R / Rfree (%) | 21.6 / 24.3 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121.
(pdb code 7poo). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121., PDB code: 7poo:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121., PDB code: 7poo:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 7poo
Go back to
Zinc binding site 1 out
of 4 in the Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121.
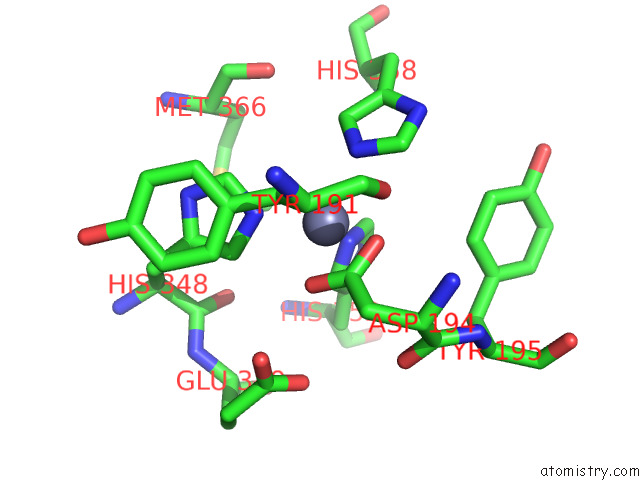
Mono view
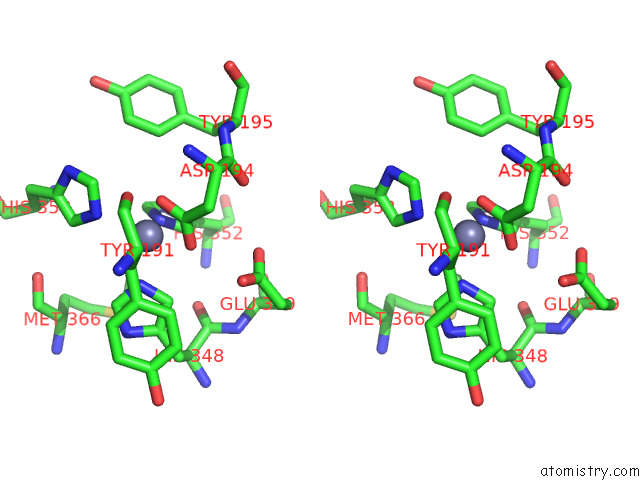
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121. within 5.0Å range:
|
Zinc binding site 2 out of 4 in 7poo
Go back to
Zinc binding site 2 out
of 4 in the Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121.
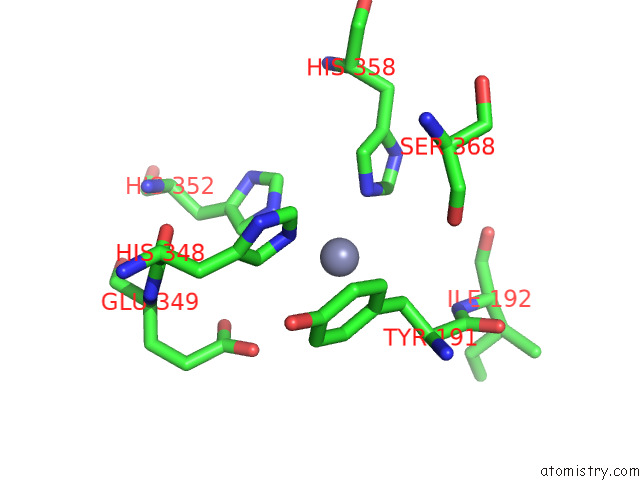
Mono view
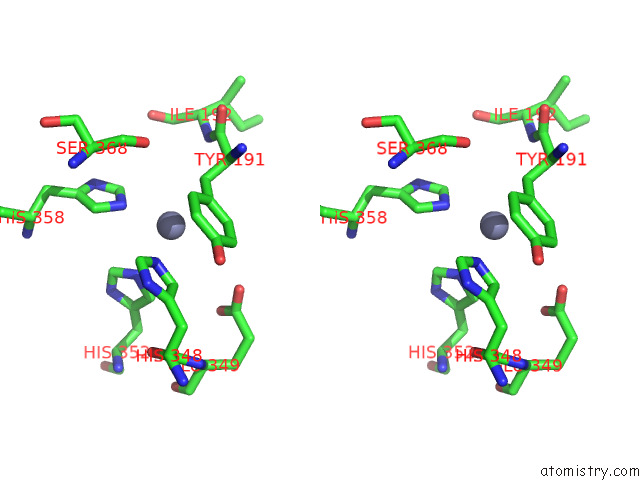
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121. within 5.0Å range:
|
Zinc binding site 3 out of 4 in 7poo
Go back to
Zinc binding site 3 out
of 4 in the Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121.
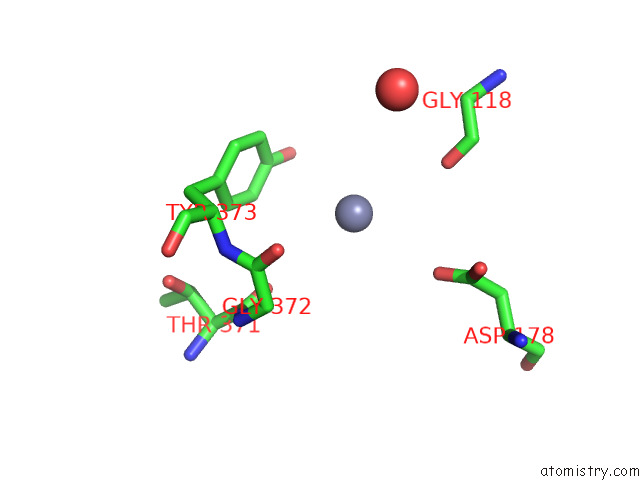
Mono view
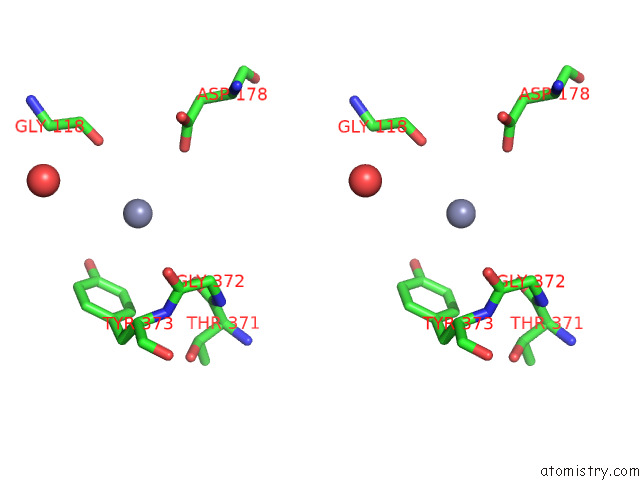
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121. within 5.0Å range:
|
Zinc binding site 4 out of 4 in 7poo
Go back to
Zinc binding site 4 out
of 4 in the Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121.
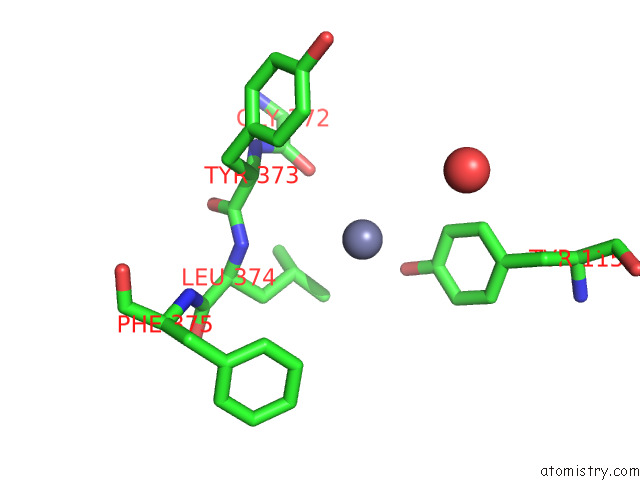
Mono view
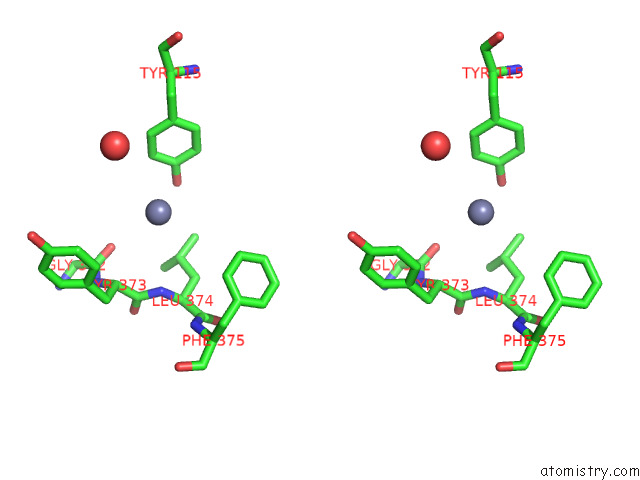
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of Profragilysin-3 (Probft-3) From Bacteroides Fragilis in Complex with Foliosidine in P212121. within 5.0Å range:
|
Reference:
A.Jimenez-Alesanco,
U.Eckhard,
M.Asencio Del Rio,
S.Vega,
T.Guevara,
A.Velazquez-Campoy,
F.X.Gomis-Ruth,
O.Abian.
Repositioning Small Molecule Drugs As Allosteric Inhibitors of the Bft-3 Toxin From Enterotoxigenic Bacteroides Fragilis. Protein Sci. V. 31 E4427 2022.
ISSN: ESSN 1469-896X
PubMed: 36173175
DOI: 10.1002/PRO.4427
Page generated: Fri Aug 22 03:29:43 2025
ISSN: ESSN 1469-896X
PubMed: 36173175
DOI: 10.1002/PRO.4427
Last articles
Zn in 8GWBZn in 8GWE
Zn in 8GW1
Zn in 8GVX
Zn in 8GVZ
Zn in 8GW0
Zn in 8GVW
Zn in 8GV3
Zn in 8GVM
Zn in 8GSQ