Zinc »
PDB 7nup-7o4f »
7nz9 »
Zinc in PDB 7nz9: Crystal Structure of Mouse ADAT2/ADAT3 Trna Deamination Complex V128L Mutant
Enzymatic activity of Crystal Structure of Mouse ADAT2/ADAT3 Trna Deamination Complex V128L Mutant
All present enzymatic activity of Crystal Structure of Mouse ADAT2/ADAT3 Trna Deamination Complex V128L Mutant:
3.5.4.33;
3.5.4.33;
Protein crystallography data
The structure of Crystal Structure of Mouse ADAT2/ADAT3 Trna Deamination Complex V128L Mutant, PDB code: 7nz9
was solved by
E.Ramos Morales,
C.Romier,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.38 / 1.99 |
| Space group | P 2 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 51.976, 106.814, 129.668, 90, 90, 90 |
| R / Rfree (%) | 18.9 / 21.8 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Mouse ADAT2/ADAT3 Trna Deamination Complex V128L Mutant
(pdb code 7nz9). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Crystal Structure of Mouse ADAT2/ADAT3 Trna Deamination Complex V128L Mutant, PDB code: 7nz9:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Crystal Structure of Mouse ADAT2/ADAT3 Trna Deamination Complex V128L Mutant, PDB code: 7nz9:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 7nz9
Go back to
Zinc binding site 1 out
of 2 in the Crystal Structure of Mouse ADAT2/ADAT3 Trna Deamination Complex V128L Mutant
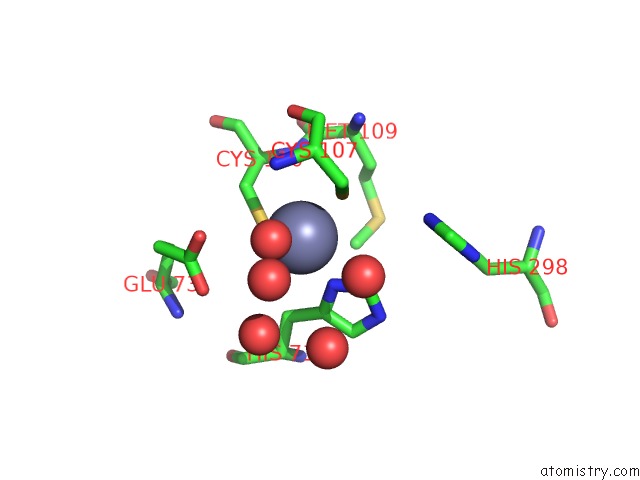
Mono view
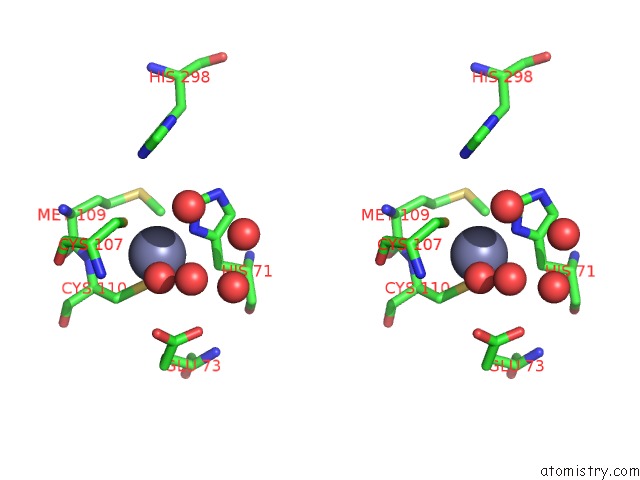
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Mouse ADAT2/ADAT3 Trna Deamination Complex V128L Mutant within 5.0Å range:
|
Zinc binding site 2 out of 2 in 7nz9
Go back to
Zinc binding site 2 out
of 2 in the Crystal Structure of Mouse ADAT2/ADAT3 Trna Deamination Complex V128L Mutant
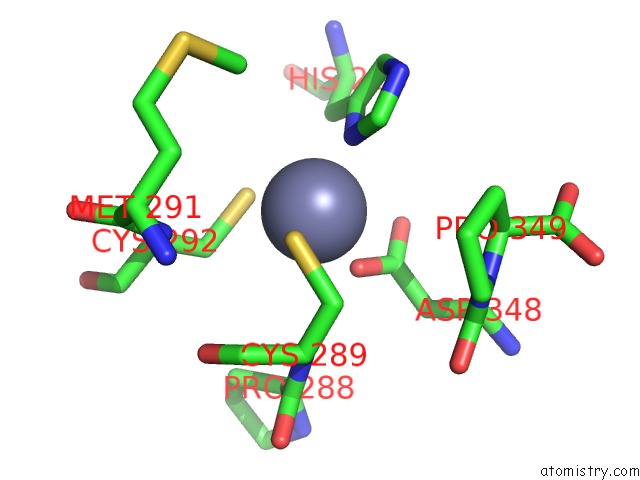
Mono view
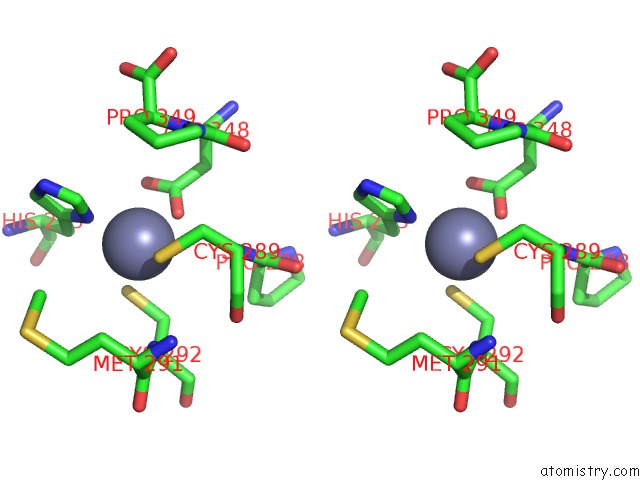
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Mouse ADAT2/ADAT3 Trna Deamination Complex V128L Mutant within 5.0Å range:
|
Reference:
E.Ramos Morales,
C.Romier.
The Structure of the Mouse ADAT2/ADAT3 Complex Reveals the Molecular Basis For Mammalian Trna Wobble Adenosine to Inosine Deamination To Be Published.
Page generated: Fri Aug 22 02:45:35 2025
Last articles
Zn in 8DZFZn in 8DZE
Zn in 8DZJ
Zn in 8DY9
Zn in 8DY7
Zn in 8DYQ
Zn in 8DWL
Zn in 8DWJ
Zn in 8DQN
Zn in 8DW6