Zinc »
PDB 7m5o-7ml4 »
7mc5 »
Zinc in PDB 7mc5: Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex
Protein crystallography data
The structure of Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex, PDB code: 7mc5
was solved by
N.M.Moeller,
K.Shi,
S.Banerjee,
L.Yin,
H.Aihara,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 57.70 / 1.64 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 63.735, 67.48, 111.249, 90, 90, 90 |
| R / Rfree (%) | 16.6 / 19.7 |
Other elements in 7mc5:
The structure of Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex also contains other interesting chemical elements:
| Chlorine | (Cl) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex
(pdb code 7mc5). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex, PDB code: 7mc5:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex, PDB code: 7mc5:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 7mc5
Go back to
Zinc binding site 1 out
of 4 in the Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex
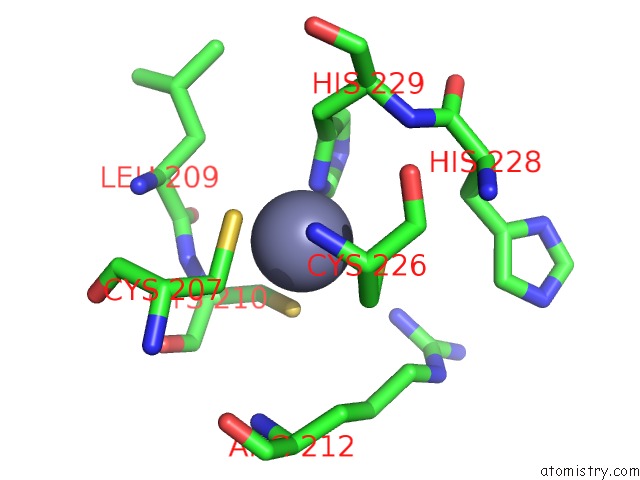
Mono view
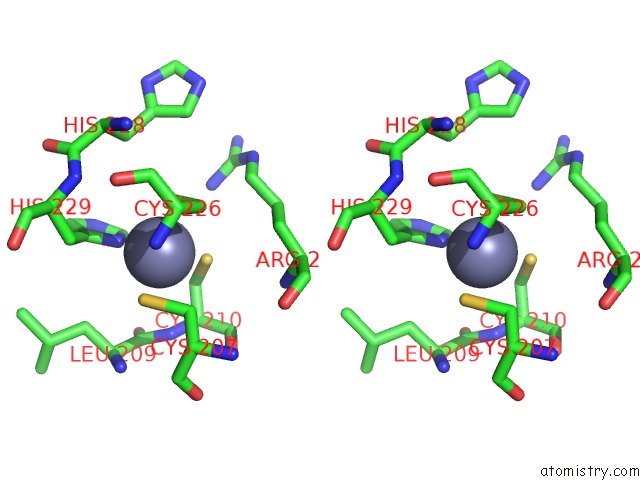
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex within 5.0Å range:
|
Zinc binding site 2 out of 4 in 7mc5
Go back to
Zinc binding site 2 out
of 4 in the Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex
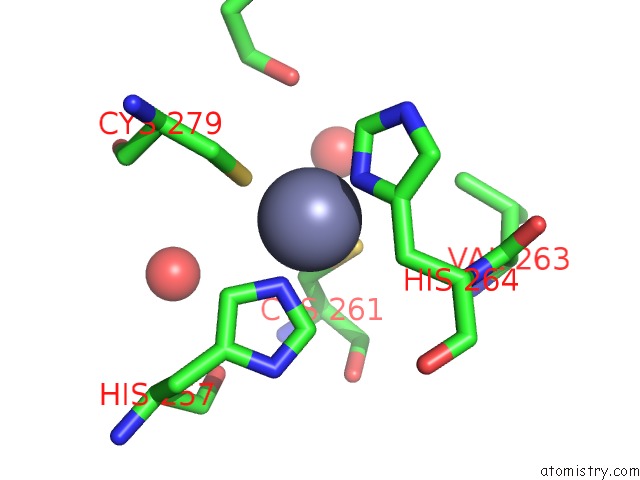
Mono view
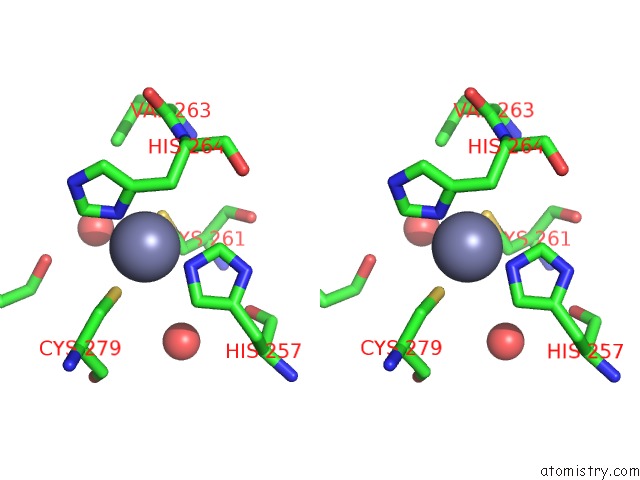
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex within 5.0Å range:
|
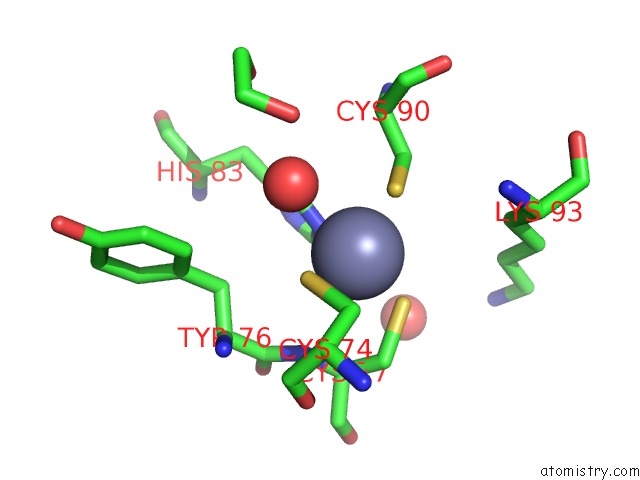
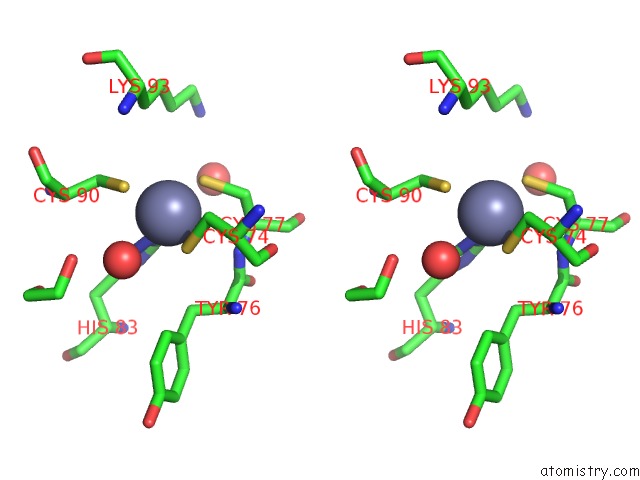
Zinc binding site 3 out of 4 in 7mc5
Go back to
Zinc binding site 3 out
of 4 in the Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex within 5.0Å range:
|
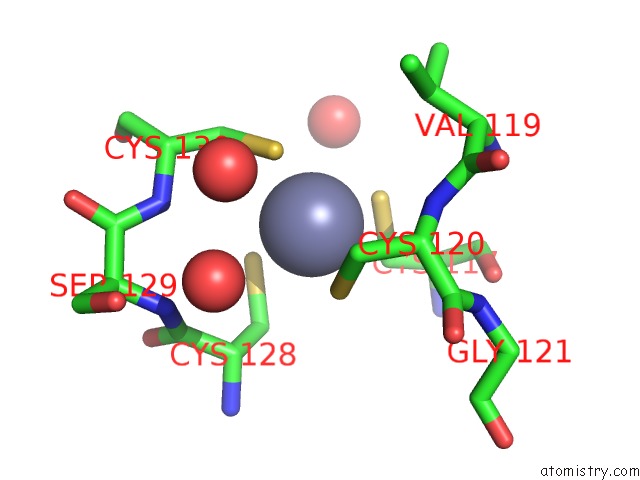
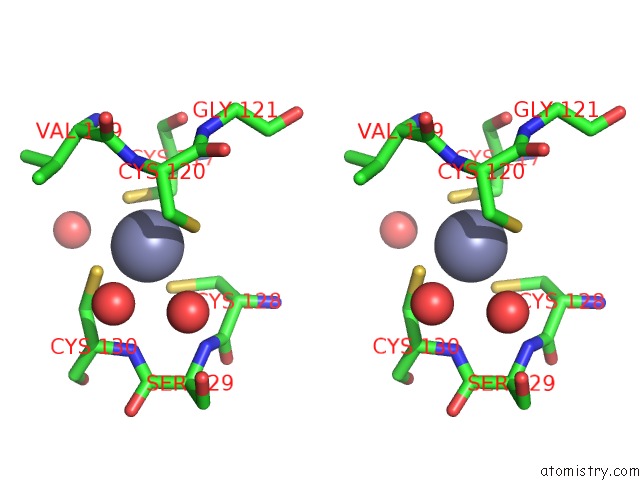
Zinc binding site 4 out of 4 in 7mc5
Go back to
Zinc binding site 4 out
of 4 in the Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of the Sars-Cov-2 Exon-NSP10 Complex within 5.0Å range:
|
Reference:
N.H.Moeller,
K.Shi,
O.Demir,
S.Banerjee,
L.Yin,
C.Belica,
C.Durfee,
R.E.Amaro,
H.Aihara.
Structure and Dynamics of Sars-Cov-2 Proofreading Exoribonuclease Exon. Biorxiv 2021.
PubMed: 33821277
DOI: 10.1101/2021.04.02.438274
Page generated: Fri Aug 22 02:05:08 2025
PubMed: 33821277
DOI: 10.1101/2021.04.02.438274
Last articles
Zn in 7WSSZn in 7WT0
Zn in 7WSZ
Zn in 7WSK
Zn in 7WSJ
Zn in 7WQG
Zn in 7WQL
Zn in 7WSH
Zn in 7WSG
Zn in 7WSF