Zinc »
PDB 7c3s-7ci4 »
7cf6 »
Zinc in PDB 7cf6: Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide
Protein crystallography data
The structure of Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide, PDB code: 7cf6
was solved by
I.Dhanasingh,
J.W.La,
D.W.Lee,
S.H.Lee,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.66 / 2.75 |
| Space group | P 2 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 78.609, 150.425, 151.996, 90, 90, 90 |
| R / Rfree (%) | 19.6 / 27.8 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide
(pdb code 7cf6). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 8 binding sites of Zinc where determined in the Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide, PDB code: 7cf6:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Zinc where determined in the Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide, PDB code: 7cf6:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Zinc binding site 1 out of 8 in 7cf6
Go back to
Zinc binding site 1 out
of 8 in the Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide
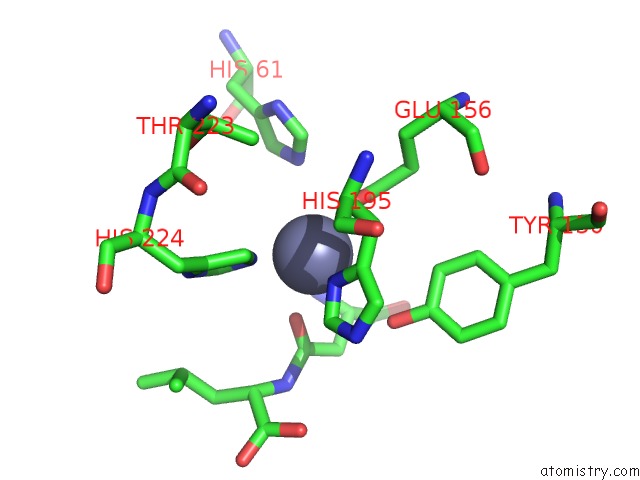
Mono view
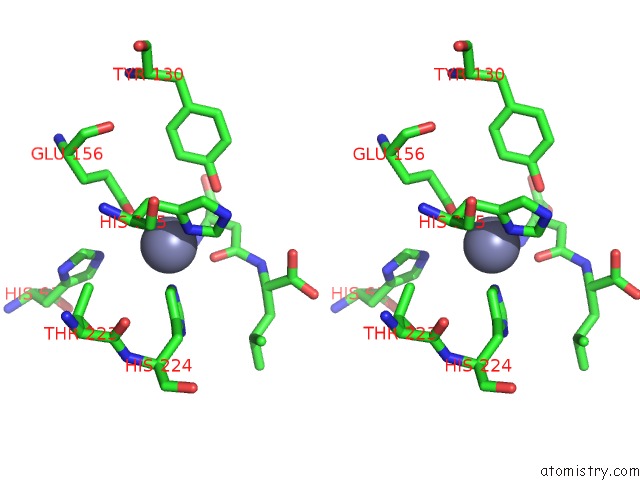
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide within 5.0Å range:
|
Zinc binding site 2 out of 8 in 7cf6
Go back to
Zinc binding site 2 out
of 8 in the Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide
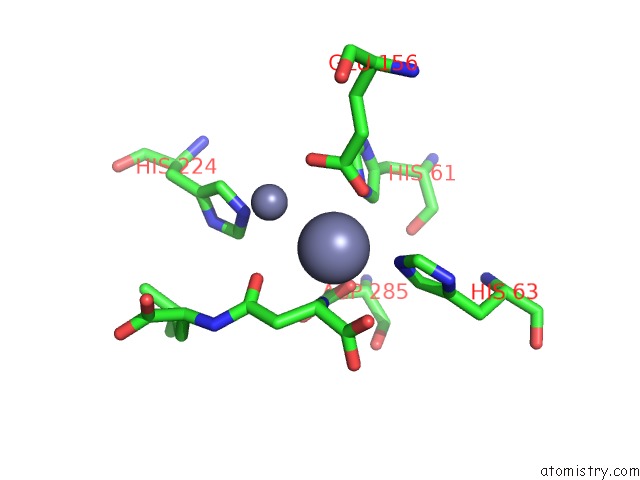
Mono view
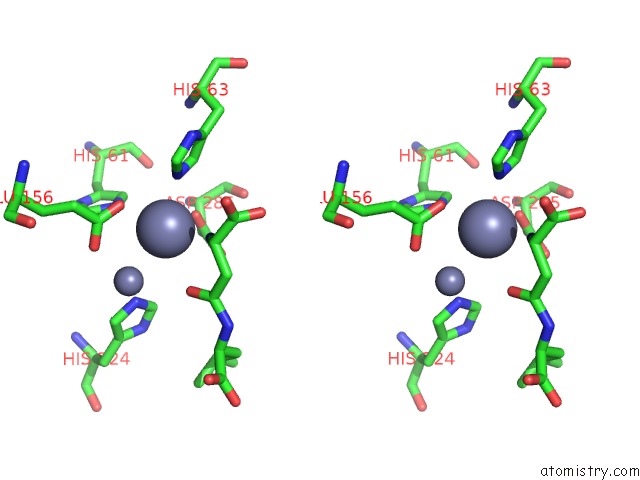
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide within 5.0Å range:
|
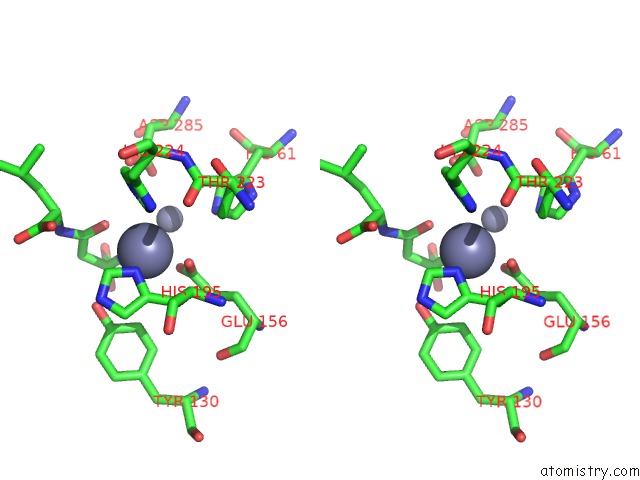
Zinc binding site 3 out of 8 in 7cf6
Go back to
Zinc binding site 3 out
of 8 in the Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide
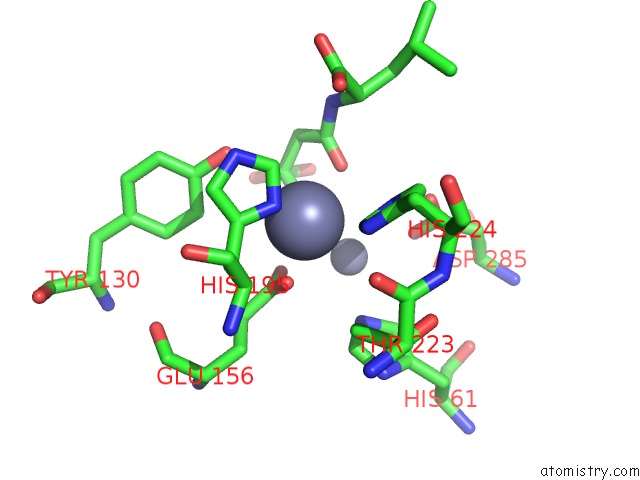
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide within 5.0Å range:
|
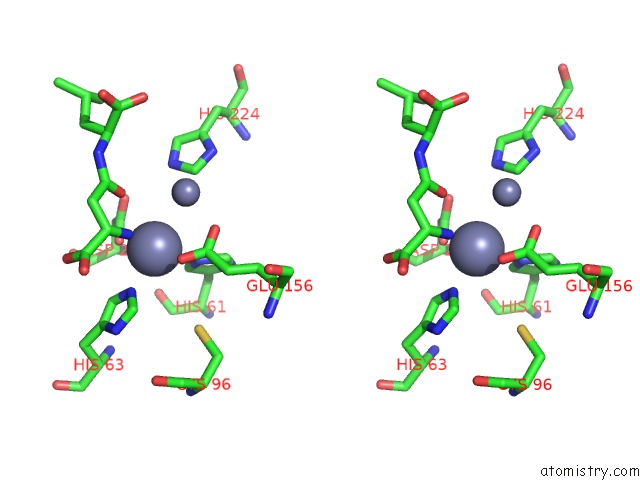
Zinc binding site 4 out of 8 in 7cf6
Go back to
Zinc binding site 4 out
of 8 in the Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide
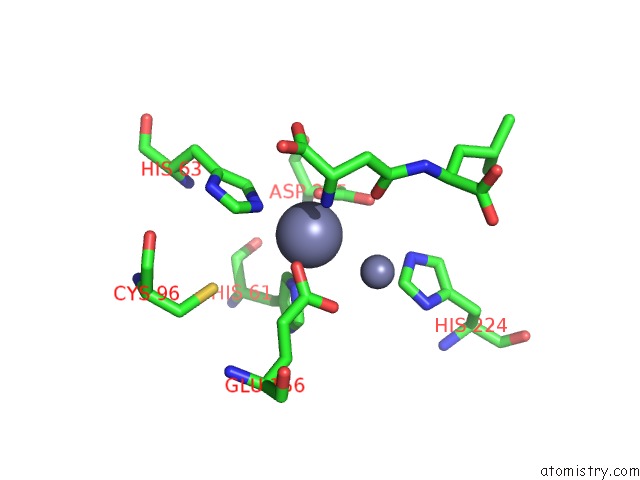
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide within 5.0Å range:
|
Zinc binding site 5 out of 8 in 7cf6
Go back to
Zinc binding site 5 out
of 8 in the Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide
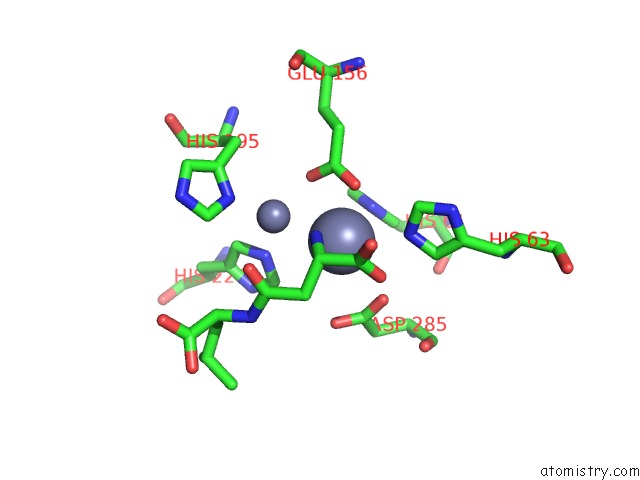
Mono view
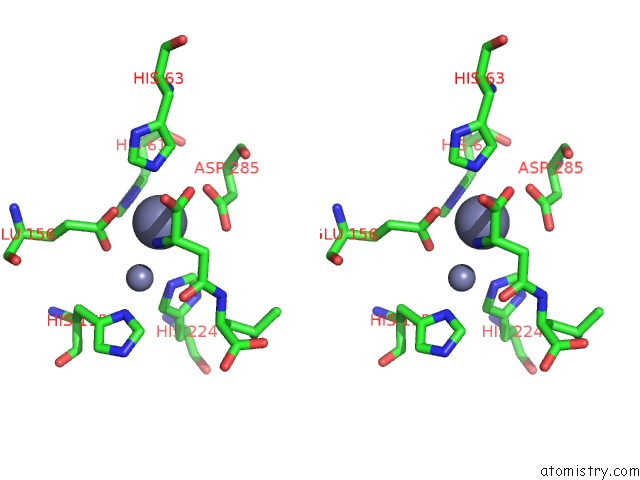
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide within 5.0Å range:
|
Zinc binding site 6 out of 8 in 7cf6
Go back to
Zinc binding site 6 out
of 8 in the Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide
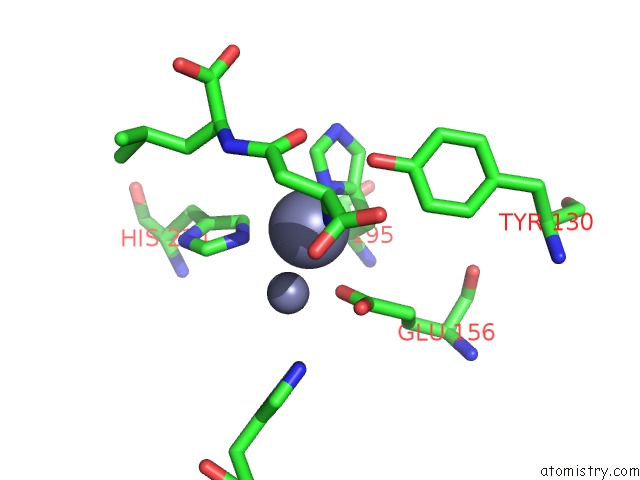
Mono view
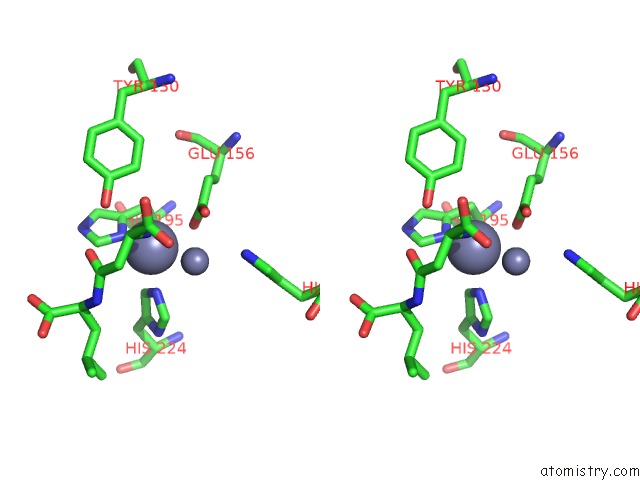
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide within 5.0Å range:
|
Zinc binding site 7 out of 8 in 7cf6
Go back to
Zinc binding site 7 out
of 8 in the Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide
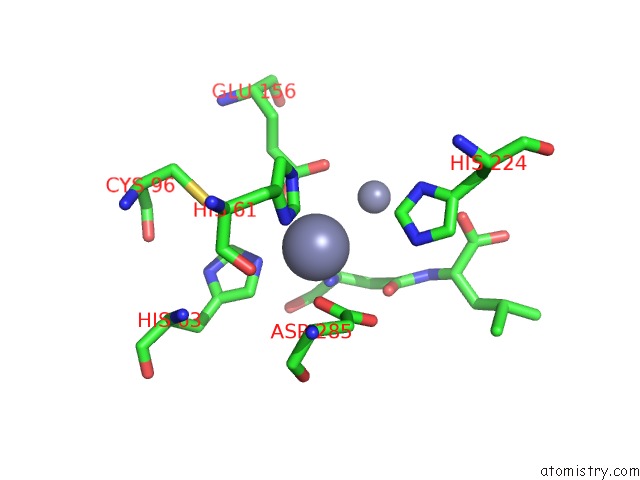
Mono view
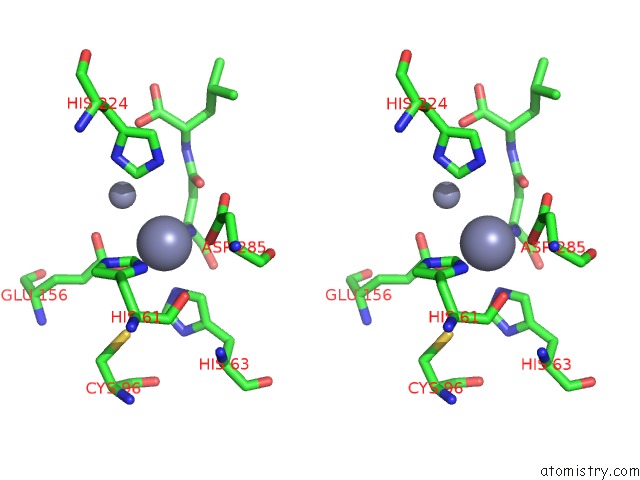
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide within 5.0Å range:
|
Zinc binding site 8 out of 8 in 7cf6
Go back to
Zinc binding site 8 out
of 8 in the Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide
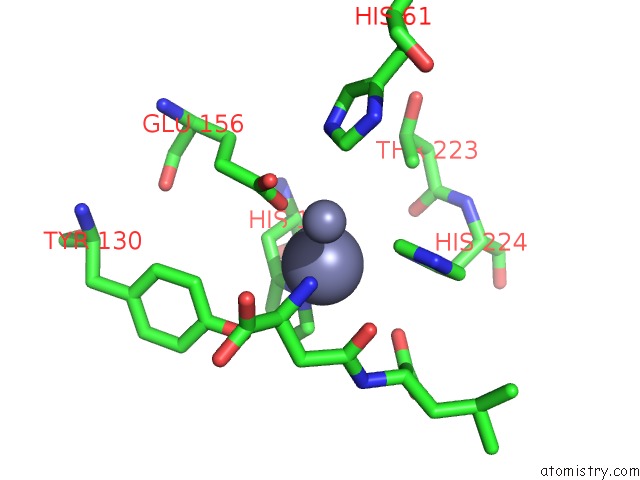
Mono view
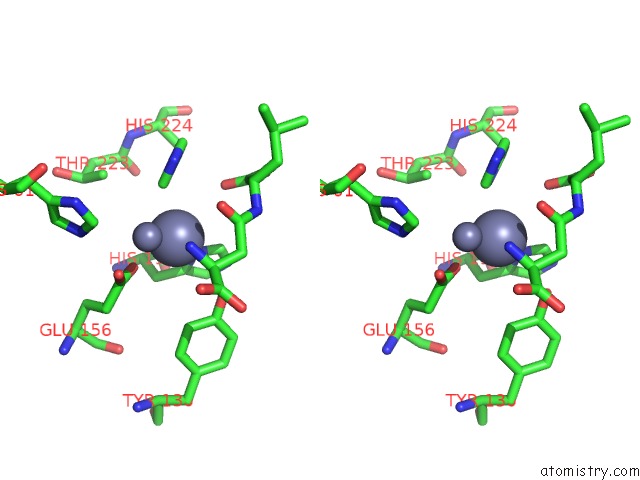
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 8 of Crystal Structure of Beta-Aspartyl Dipeptidase From Thermophilic Keratin Degrading Fervidobacterium Islandicum Aw-1 in Complex with Beta-Asp-Leu Dipeptide within 5.0Å range:
|
Reference:
J.W.La,
I.Dhanasingh,
H.Jang,
S.H.Lee,
D.W.Lee.
Functional Characterization of Primordial Protein Repair Enzyme M38 Metallo-Peptidase From Fervidobacterium Islandicum Aw-1. Front Mol Biosci V. 7 00634 2020.
ISSN: ESSN 2296-889X
PubMed: 33392259
DOI: 10.3389/FMOLB.2020.600634
Page generated: Tue Oct 29 18:11:41 2024
ISSN: ESSN 2296-889X
PubMed: 33392259
DOI: 10.3389/FMOLB.2020.600634
Last articles
Al in 3WGUAl in 3SYN
Al in 3W6P
Al in 3T34
Al in 3UKD
Al in 3RFU
Al in 3T12
Al in 3RUW
Al in 3SR0
Al in 3SS8