Zinc »
PDB 7av2-7b93 »
7b4f »
Zinc in PDB 7b4f: Structural Basis of Reactivation of Oncogenic P53 Mutants By A Small Molecule: Methylene Quinuclidinone (Mq). Human P53DBD-R282W Mutant Bound to Dna: R282W-Mq (I)
Protein crystallography data
The structure of Structural Basis of Reactivation of Oncogenic P53 Mutants By A Small Molecule: Methylene Quinuclidinone (Mq). Human P53DBD-R282W Mutant Bound to Dna: R282W-Mq (I), PDB code: 7b4f
was solved by
H.Rozenberg,
O.Degtjarik,
Z.Shakked,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 18.42 / 1.78 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 137.21, 49.743, 33.816, 90, 93.36, 90 |
| R / Rfree (%) | 15.5 / 19.5 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structural Basis of Reactivation of Oncogenic P53 Mutants By A Small Molecule: Methylene Quinuclidinone (Mq). Human P53DBD-R282W Mutant Bound to Dna: R282W-Mq (I)
(pdb code 7b4f). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Structural Basis of Reactivation of Oncogenic P53 Mutants By A Small Molecule: Methylene Quinuclidinone (Mq). Human P53DBD-R282W Mutant Bound to Dna: R282W-Mq (I), PDB code: 7b4f:
In total only one binding site of Zinc was determined in the Structural Basis of Reactivation of Oncogenic P53 Mutants By A Small Molecule: Methylene Quinuclidinone (Mq). Human P53DBD-R282W Mutant Bound to Dna: R282W-Mq (I), PDB code: 7b4f:
Zinc binding site 1 out of 1 in 7b4f
Go back to
Zinc binding site 1 out
of 1 in the Structural Basis of Reactivation of Oncogenic P53 Mutants By A Small Molecule: Methylene Quinuclidinone (Mq). Human P53DBD-R282W Mutant Bound to Dna: R282W-Mq (I)
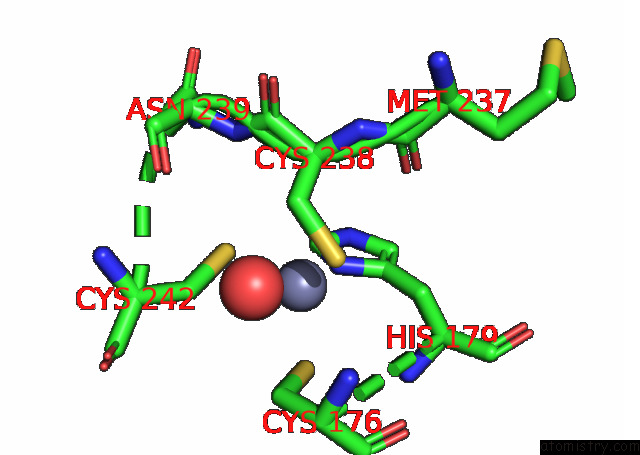
Mono view
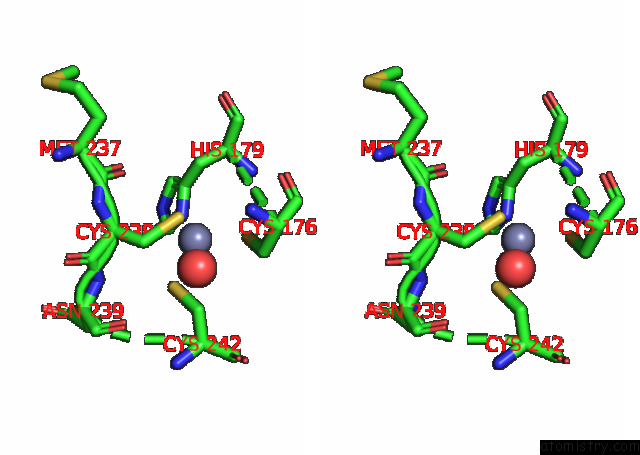
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structural Basis of Reactivation of Oncogenic P53 Mutants By A Small Molecule: Methylene Quinuclidinone (Mq). Human P53DBD-R282W Mutant Bound to Dna: R282W-Mq (I) within 5.0Å range:
|
Reference:
O.Degtjarik,
D.Golovenko,
Y.Diskin-Posner,
L.Abrahmsen,
H.Rozenberg,
Z.Shakked.
P53 Structure 9 To Be Published.
Page generated: Tue Oct 29 17:26:15 2024
Last articles
Al in 3N5KAl in 3PUW
Al in 3QPW
Al in 3LVR
Al in 3HB1
Al in 3KQL
Al in 3CMX
Al in 3LVQ
Al in 3I62
Al in 3EX7