Zinc »
PDB 6rwo-6s6b »
6ryr »
Zinc in PDB 6ryr: Nucleosome-CHD4 Complex Structure (Single CHD4 Copy)
Enzymatic activity of Nucleosome-CHD4 Complex Structure (Single CHD4 Copy)
All present enzymatic activity of Nucleosome-CHD4 Complex Structure (Single CHD4 Copy):
3.6.4.12;
3.6.4.12;
Other elements in 6ryr:
The structure of Nucleosome-CHD4 Complex Structure (Single CHD4 Copy) also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the Nucleosome-CHD4 Complex Structure (Single CHD4 Copy)
(pdb code 6ryr). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Nucleosome-CHD4 Complex Structure (Single CHD4 Copy), PDB code: 6ryr:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Nucleosome-CHD4 Complex Structure (Single CHD4 Copy), PDB code: 6ryr:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 6ryr
Go back to
Zinc binding site 1 out
of 2 in the Nucleosome-CHD4 Complex Structure (Single CHD4 Copy)

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Nucleosome-CHD4 Complex Structure (Single CHD4 Copy) within 5.0Å range:
|
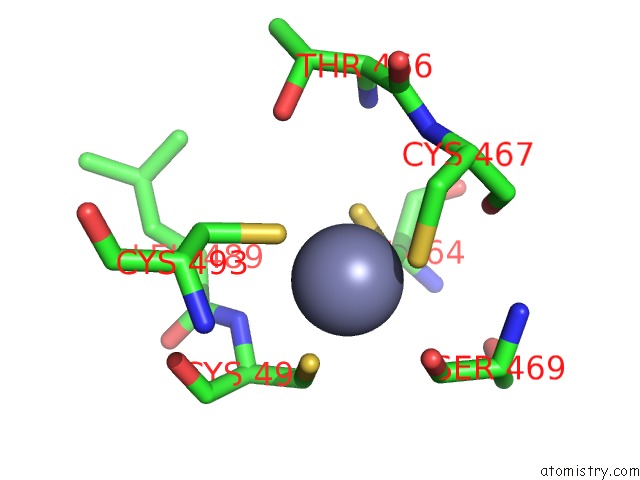
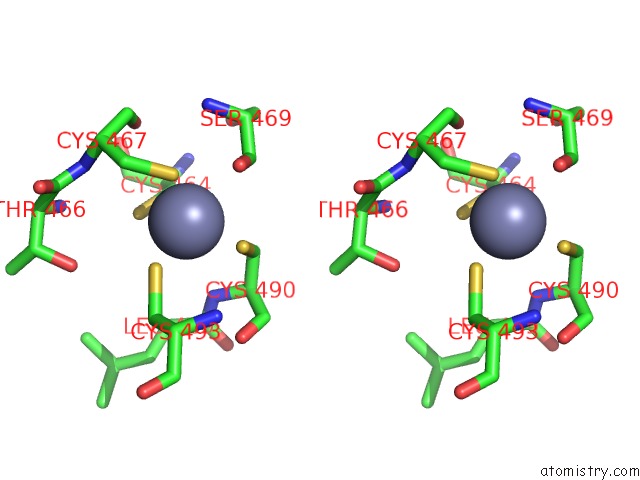
Zinc binding site 2 out of 2 in 6ryr
Go back to
Zinc binding site 2 out
of 2 in the Nucleosome-CHD4 Complex Structure (Single CHD4 Copy)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Nucleosome-CHD4 Complex Structure (Single CHD4 Copy) within 5.0Å range:
|
Reference:
L.Farnung,
M.Ochmann,
P.Cramer.
Nucleosome-CHD4 Chromatin Remodeller Structure Maps Human Disease Mutations. Elife V. 9 2020.
ISSN: ESSN 2050-084X
PubMed: 32543371
DOI: 10.7554/ELIFE.56178
Page generated: Tue Oct 29 06:55:54 2024
ISSN: ESSN 2050-084X
PubMed: 32543371
DOI: 10.7554/ELIFE.56178
Last articles
Al in 7BILAl in 6KS6
Al in 7BSS
Al in 7BSQ
Al in 6ZHG
Al in 6XV0
Al in 6T66
Al in 6XMS
Al in 6XEZ
Al in 6XI8