Zinc »
PDB 6nl8-6o5g »
6ny6 »
Zinc in PDB 6ny6: Structure of Dimeric Escherichia Coli Toxin Yoeb Bound to the Thermus Thermophilus 30S Ribosome
Protein crystallography data
The structure of Structure of Dimeric Escherichia Coli Toxin Yoeb Bound to the Thermus Thermophilus 30S Ribosome, PDB code: 6ny6
was solved by
I.J.Pavelich,
E.D.Hoffer,
T.Maehigashi,
C.M.Dunham,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 49.83 / 3.74 |
| Space group | P 41 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 401.710, 401.710, 175.440, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 20.6 / 22.4 |
Other elements in 6ny6:
The structure of Structure of Dimeric Escherichia Coli Toxin Yoeb Bound to the Thermus Thermophilus 30S Ribosome also contains other interesting chemical elements:
| Magnesium | (Mg) | 184 atoms |
| Iron | (Fe) | 4 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of Dimeric Escherichia Coli Toxin Yoeb Bound to the Thermus Thermophilus 30S Ribosome
(pdb code 6ny6). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Structure of Dimeric Escherichia Coli Toxin Yoeb Bound to the Thermus Thermophilus 30S Ribosome, PDB code: 6ny6:
In total only one binding site of Zinc was determined in the Structure of Dimeric Escherichia Coli Toxin Yoeb Bound to the Thermus Thermophilus 30S Ribosome, PDB code: 6ny6:
Zinc binding site 1 out of 1 in 6ny6
Go back to
Zinc binding site 1 out
of 1 in the Structure of Dimeric Escherichia Coli Toxin Yoeb Bound to the Thermus Thermophilus 30S Ribosome
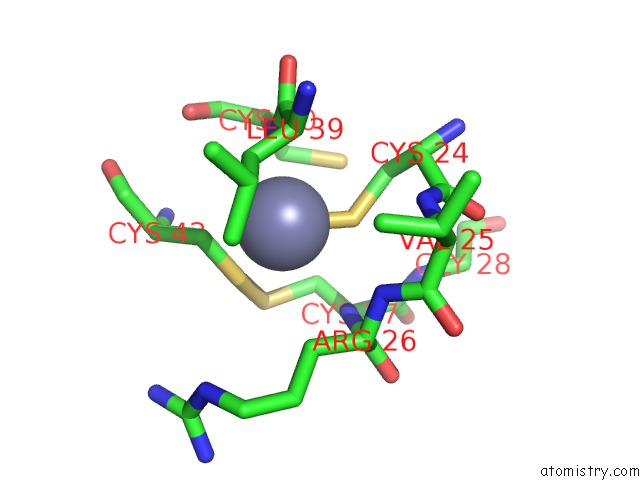
Mono view
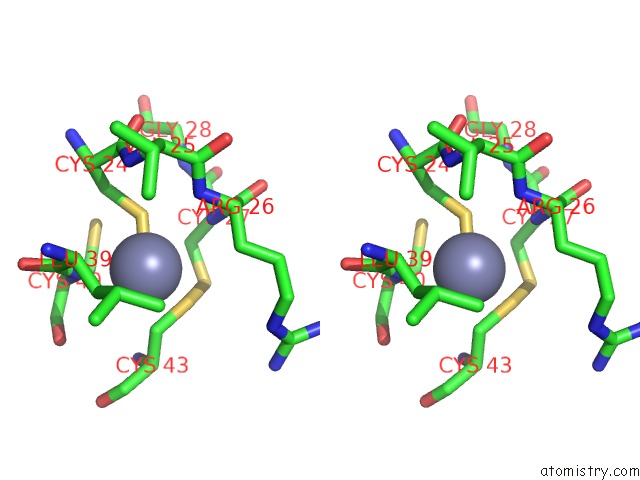
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of Dimeric Escherichia Coli Toxin Yoeb Bound to the Thermus Thermophilus 30S Ribosome within 5.0Å range:
|
Reference:
I.J.Pavelich,
T.Maehigashi,
E.D.Hoffer,
A.Ruangprasert,
S.J.Miles,
C.M.Dunham.
Monomeric Yoeb Toxin Retains Rnase Activity But Adopts An Obligate Dimeric Form For Thermal Stability. Nucleic Acids Res. V. 47 10400 2019.
ISSN: ESSN 1362-4962
PubMed: 31501867
DOI: 10.1093/NAR/GKZ760
Page generated: Tue Oct 29 04:01:22 2024
ISSN: ESSN 1362-4962
PubMed: 31501867
DOI: 10.1093/NAR/GKZ760
Last articles
As in 3N68As in 3N67
As in 3N6A
As in 3N69
As in 3N5T
As in 3N5S
As in 3N5R
As in 3M1R
As in 3N5Q
As in 3N5P