Zinc »
PDB 6ngn-6nla »
6nj5 »
Zinc in PDB 6nj5: Thermostable Variant of Human Carbonic Anhydrase II with Disordered Tetrazine 2.0 at Site 233
Enzymatic activity of Thermostable Variant of Human Carbonic Anhydrase II with Disordered Tetrazine 2.0 at Site 233
All present enzymatic activity of Thermostable Variant of Human Carbonic Anhydrase II with Disordered Tetrazine 2.0 at Site 233:
4.2.1.1;
4.2.1.1;
Protein crystallography data
The structure of Thermostable Variant of Human Carbonic Anhydrase II with Disordered Tetrazine 2.0 at Site 233, PDB code: 6nj5
was solved by
K.M.Kean,
P.A.Karplus,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 37.88 / 1.25 |
| Space group | C 2 2 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 44.419, 72.460, 151.508, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.6 / 20.6 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Thermostable Variant of Human Carbonic Anhydrase II with Disordered Tetrazine 2.0 at Site 233
(pdb code 6nj5). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Thermostable Variant of Human Carbonic Anhydrase II with Disordered Tetrazine 2.0 at Site 233, PDB code: 6nj5:
In total only one binding site of Zinc was determined in the Thermostable Variant of Human Carbonic Anhydrase II with Disordered Tetrazine 2.0 at Site 233, PDB code: 6nj5:
Zinc binding site 1 out of 1 in 6nj5
Go back to
Zinc binding site 1 out
of 1 in the Thermostable Variant of Human Carbonic Anhydrase II with Disordered Tetrazine 2.0 at Site 233
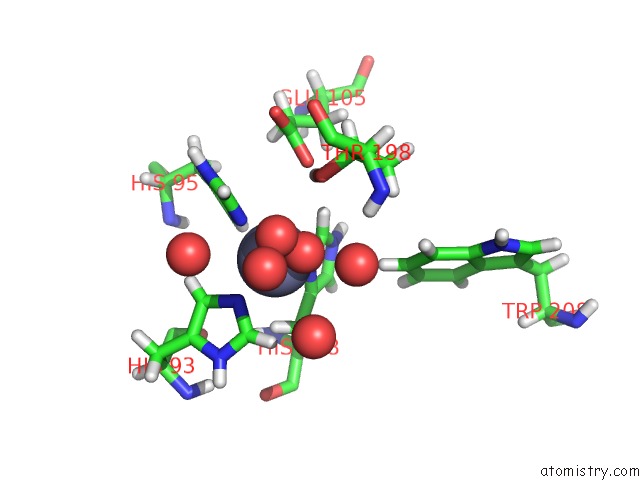
Mono view
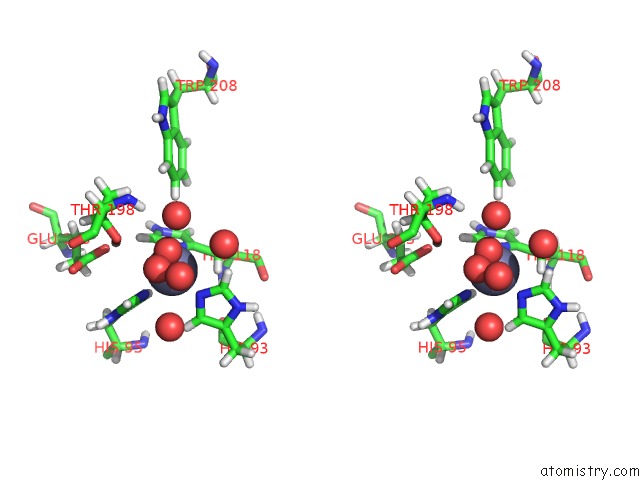
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Thermostable Variant of Human Carbonic Anhydrase II with Disordered Tetrazine 2.0 at Site 233 within 5.0Å range:
|
Reference:
K.M.Kean,
N.G.Waugh,
R.Bednar,
P.A.Karplus,
R.A.Mehl.
Genetic Code Expansion of Thermostable Carbonic Anhydrase II to Create Ideal Platform For Bioorthogonal Ligating Industrial Useful Enzymes to Synthetic Polymers To Be Published.
Page generated: Thu Aug 21 17:49:45 2025
Last articles
Zn in 6XEBZn in 6XDM
Zn in 6XD1
Zn in 6XCI
Zn in 6XD0
Zn in 6XCF
Zn in 6XCD
Zn in 6XCC
Zn in 6XCB
Zn in 6XB8