Zinc »
PDB 6knc-6kua »
6knc »
Zinc in PDB 6knc: Pold-Pcna-Dna (Form B)
Enzymatic activity of Pold-Pcna-Dna (Form B)
Other elements in 6knc:
The structure of Pold-Pcna-Dna (Form B) also contains other interesting chemical elements:
| Iron | (Fe) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the Pold-Pcna-Dna (Form B)
(pdb code 6knc). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Pold-Pcna-Dna (Form B), PDB code: 6knc:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Pold-Pcna-Dna (Form B), PDB code: 6knc:
Jump to Zinc binding site number: 1; 2; 3; 4;
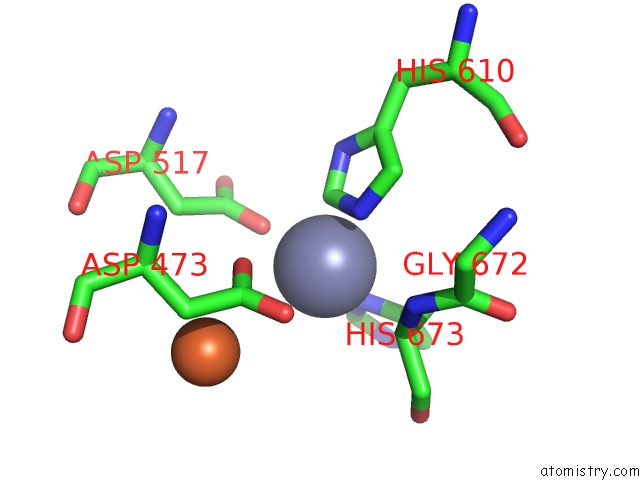
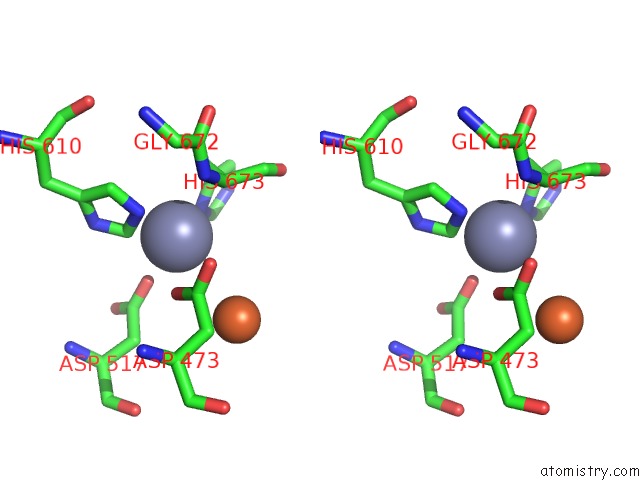
Zinc binding site 1 out of 4 in 6knc
Go back to
Zinc binding site 1 out
of 4 in the Pold-Pcna-Dna (Form B)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Pold-Pcna-Dna (Form B) within 5.0Å range:
|
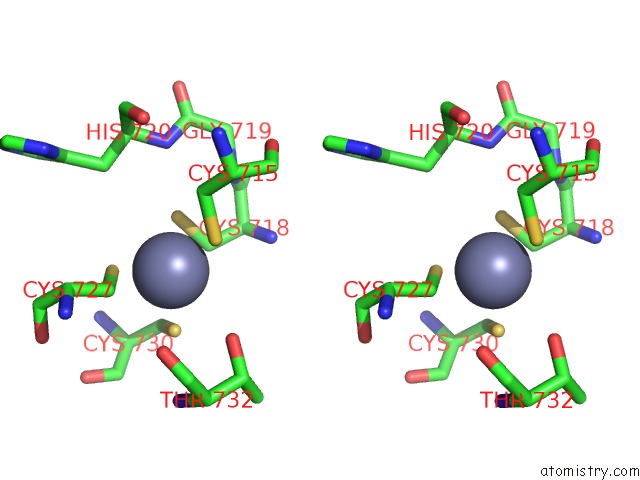
Zinc binding site 2 out of 4 in 6knc
Go back to
Zinc binding site 2 out
of 4 in the Pold-Pcna-Dna (Form B)

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Pold-Pcna-Dna (Form B) within 5.0Å range:
|
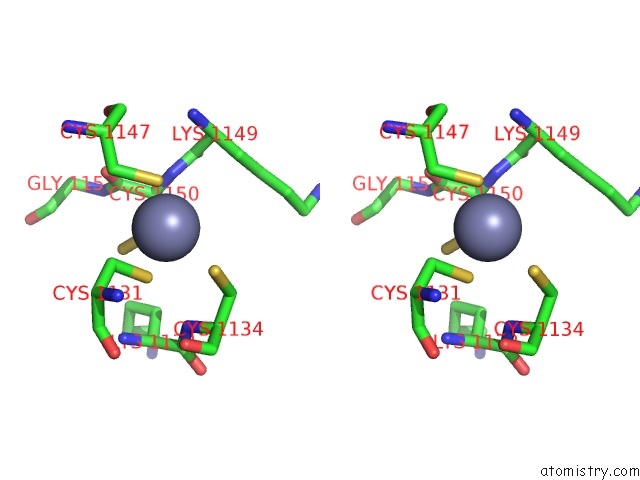
Zinc binding site 3 out of 4 in 6knc
Go back to
Zinc binding site 3 out
of 4 in the Pold-Pcna-Dna (Form B)

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Pold-Pcna-Dna (Form B) within 5.0Å range:
|
Zinc binding site 4 out of 4 in 6knc
Go back to
Zinc binding site 4 out
of 4 in the Pold-Pcna-Dna (Form B)

Mono view
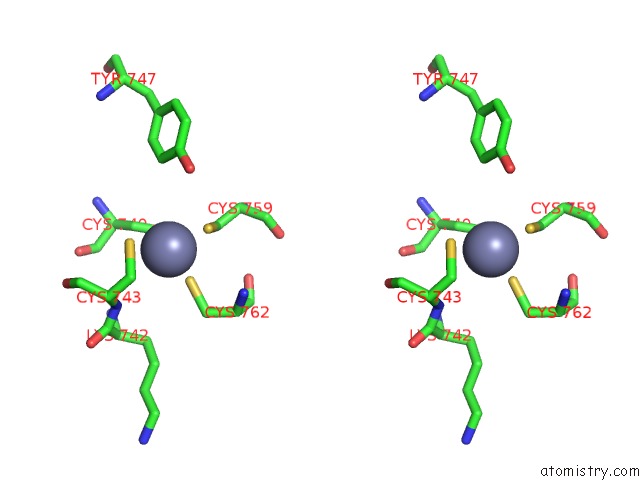
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Pold-Pcna-Dna (Form B) within 5.0Å range:
|
Reference:
K.Mayanagi,
Y.Ishino.
Molecular Architecture and Switching Mechanism of Dna Polymerase D in Replisome As Revealed By Cryo-Electron Microscopy To Be Published.
Page generated: Thu Aug 21 16:46:47 2025
Last articles
Zn in 6SJ0Zn in 6SJ3
Zn in 6SJ2
Zn in 6SJ1
Zn in 6SEV
Zn in 6SIZ
Zn in 6SIY
Zn in 6SIX
Zn in 6SIW
Zn in 6SIV