Zinc »
PDB 6btn-6c6u »
6bwy »
Zinc in PDB 6bwy: Dna Substrate Selection By APOBEC3G
Protein crystallography data
The structure of Dna Substrate Selection By APOBEC3G, PDB code: 6bwy
was solved by
S.J.Ziegler,
O.Buzovetsky,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 44.19 / 2.90 |
| Space group | P 43 |
| Cell size a, b, c (Å), α, β, γ (°) | 79.072, 79.072, 266.411, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.4 / 28.7 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Dna Substrate Selection By APOBEC3G
(pdb code 6bwy). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Dna Substrate Selection By APOBEC3G, PDB code: 6bwy:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Dna Substrate Selection By APOBEC3G, PDB code: 6bwy:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 6bwy
Go back to
Zinc binding site 1 out
of 4 in the Dna Substrate Selection By APOBEC3G

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Dna Substrate Selection By APOBEC3G within 5.0Å range:
|
Zinc binding site 2 out of 4 in 6bwy
Go back to
Zinc binding site 2 out
of 4 in the Dna Substrate Selection By APOBEC3G
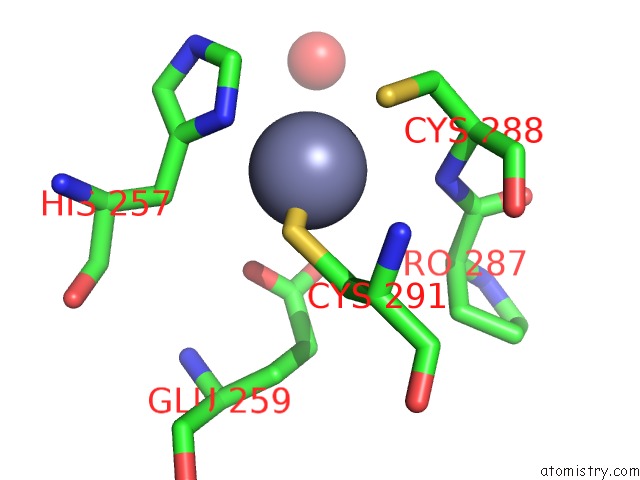
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Dna Substrate Selection By APOBEC3G within 5.0Å range:
|
Zinc binding site 3 out of 4 in 6bwy
Go back to
Zinc binding site 3 out
of 4 in the Dna Substrate Selection By APOBEC3G
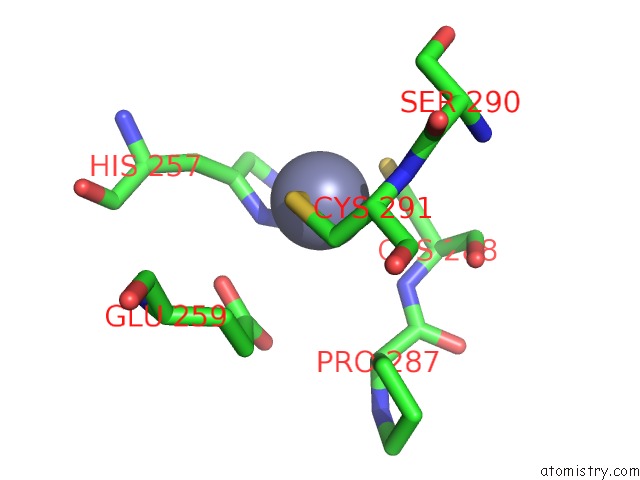
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Dna Substrate Selection By APOBEC3G within 5.0Å range:
|
Zinc binding site 4 out of 4 in 6bwy
Go back to
Zinc binding site 4 out
of 4 in the Dna Substrate Selection By APOBEC3G

Mono view
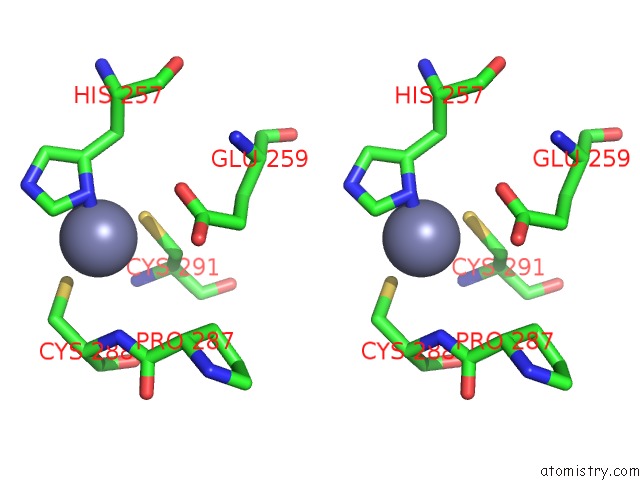
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Dna Substrate Selection By APOBEC3G within 5.0Å range:
|
Reference:
S.J.Ziegler,
C.Liu,
M.Landau,
O.Buzovetsky,
B.A.Desimmie,
Q.Zhao,
T.Sasaki,
R.C.Burdick,
V.K.Pathak,
K.S.Anderson,
Y.Xiong.
Insights Into Dna Substrate Selection By APOBEC3G From Structural, Biochemical, and Functional Studies. Plos One V. 13 95048 2018.
ISSN: ESSN 1932-6203
PubMed: 29596531
DOI: 10.1371/JOURNAL.PONE.0195048
Page generated: Mon Oct 28 18:23:18 2024
ISSN: ESSN 1932-6203
PubMed: 29596531
DOI: 10.1371/JOURNAL.PONE.0195048
Last articles
As in 3O56As in 3NVV
As in 3NSE
As in 3NLU
As in 3NLT
As in 3NRF
As in 3NLI
As in 3NLG
As in 3NLH
As in 3NLF