Zinc »
PDB 6av3-6be6 »
6b6h »
Zinc in PDB 6b6h: The Cryo-Em Structure of A Bacterial Class I Transcription Activation Complex
Enzymatic activity of The Cryo-Em Structure of A Bacterial Class I Transcription Activation Complex
All present enzymatic activity of The Cryo-Em Structure of A Bacterial Class I Transcription Activation Complex:
2.7.7.6;
2.7.7.6;
Other elements in 6b6h:
The structure of The Cryo-Em Structure of A Bacterial Class I Transcription Activation Complex also contains other interesting chemical elements:
| Magnesium | (Mg) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the The Cryo-Em Structure of A Bacterial Class I Transcription Activation Complex
(pdb code 6b6h). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the The Cryo-Em Structure of A Bacterial Class I Transcription Activation Complex, PDB code: 6b6h:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the The Cryo-Em Structure of A Bacterial Class I Transcription Activation Complex, PDB code: 6b6h:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 6b6h
Go back to
Zinc binding site 1 out
of 2 in the The Cryo-Em Structure of A Bacterial Class I Transcription Activation Complex

Mono view
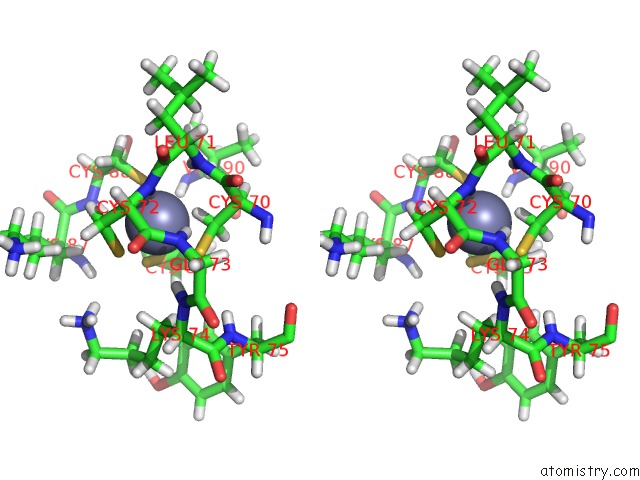
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of The Cryo-Em Structure of A Bacterial Class I Transcription Activation Complex within 5.0Å range:
|
Zinc binding site 2 out of 2 in 6b6h
Go back to
Zinc binding site 2 out
of 2 in the The Cryo-Em Structure of A Bacterial Class I Transcription Activation Complex
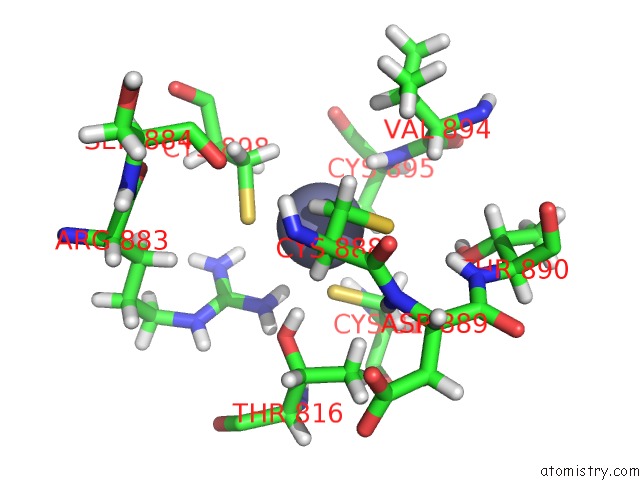
Mono view
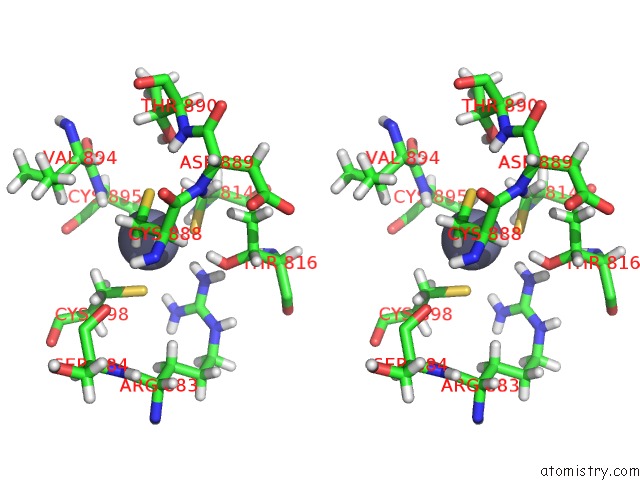
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of The Cryo-Em Structure of A Bacterial Class I Transcription Activation Complex within 5.0Å range:
|
Reference:
B.Liu,
C.Hong,
R.K.Huang,
Z.Yu,
T.A.Steitz.
Structural Basis of Bacterial Transcription Activation. Science V. 358 947 2017.
ISSN: ESSN 1095-9203
PubMed: 29146813
DOI: 10.1126/SCIENCE.AAO1923
Page generated: Mon Oct 28 17:51:19 2024
ISSN: ESSN 1095-9203
PubMed: 29146813
DOI: 10.1126/SCIENCE.AAO1923
Last articles
Al in 3CF1Al in 3C7K
Al in 3B9R
Al in 3AB3
Al in 3BH7
Al in 3AR8
Al in 2YNM
Al in 2ZJY
Al in 2ZBG
Al in 2ZBD