Zinc »
PDB 5hp2-5i3l »
5hvq »
Zinc in PDB 5hvq: Alternative Model of the Mage-G1 Nse-1 Complex
Protein crystallography data
The structure of Alternative Model of the Mage-G1 Nse-1 Complex, PDB code: 5hvq
was solved by
J.A.Newman,
C.D.O.Cooper,
A.K.Roos,
H.Aitkenhead,
U.C.T.Oppermann,
H.J.Cho,
R.Osman,
O.Gileadi,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 32.36 / 2.92 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 83.257, 154.327, 53.726, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 23.7 / 26.8 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Alternative Model of the Mage-G1 Nse-1 Complex
(pdb code 5hvq). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Alternative Model of the Mage-G1 Nse-1 Complex, PDB code: 5hvq:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Alternative Model of the Mage-G1 Nse-1 Complex, PDB code: 5hvq:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 5hvq
Go back to
Zinc binding site 1 out
of 2 in the Alternative Model of the Mage-G1 Nse-1 Complex

Mono view
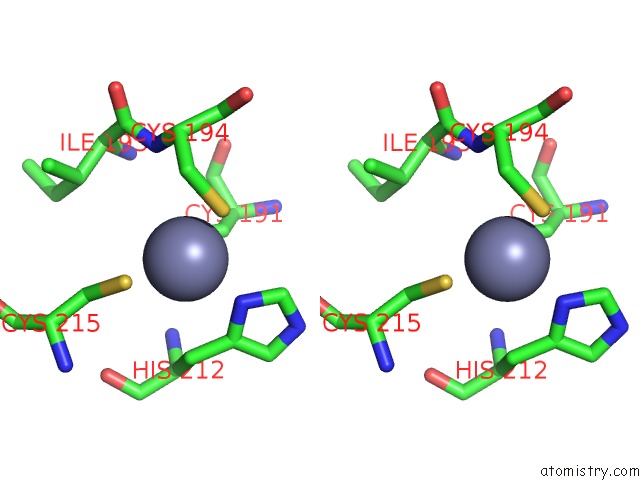
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Alternative Model of the Mage-G1 Nse-1 Complex within 5.0Å range:
|
Zinc binding site 2 out of 2 in 5hvq
Go back to
Zinc binding site 2 out
of 2 in the Alternative Model of the Mage-G1 Nse-1 Complex
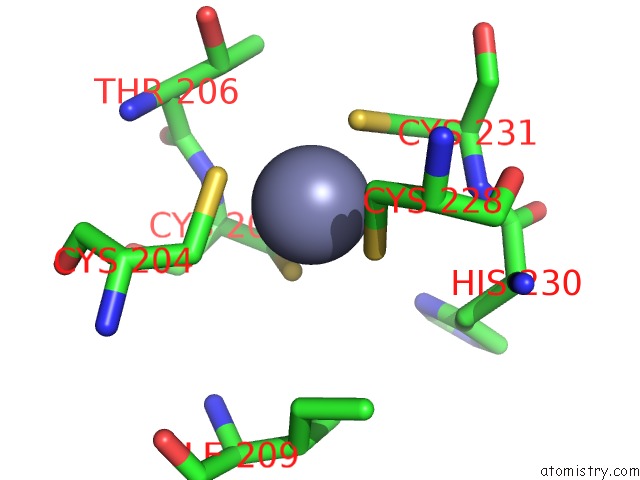
Mono view
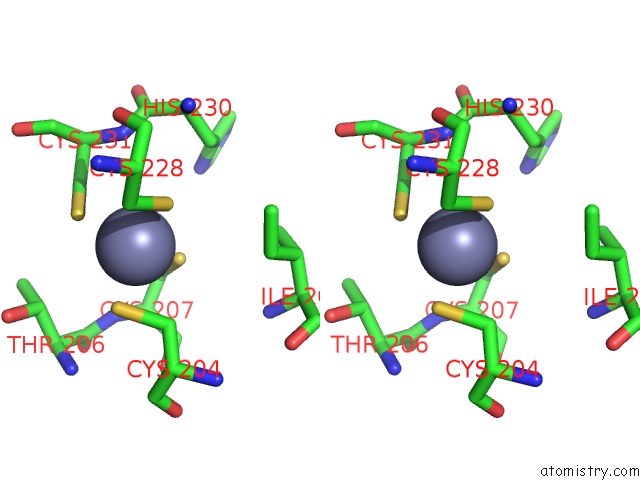
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Alternative Model of the Mage-G1 Nse-1 Complex within 5.0Å range:
|
Reference:
J.A.Newman,
C.D.Cooper,
A.K.Roos,
H.Aitkenhead,
U.C.Oppermann,
H.J.Cho,
R.Osman,
O.Gileadi.
Structures of Two Melanoma-Associated Antigens Suggest Allosteric Regulation of Effector Binding. Plos One V. 11 48762 2016.
ISSN: ESSN 1932-6203
PubMed: 26910052
DOI: 10.1371/JOURNAL.PONE.0148762
Page generated: Sun Oct 27 17:41:16 2024
ISSN: ESSN 1932-6203
PubMed: 26910052
DOI: 10.1371/JOURNAL.PONE.0148762
Last articles
Ag in 7SLMAg in 7SLK
Ag in 7SLL
Ag in 7SLJ
Ag in 7SLF
Ag in 7SLE
Ag in 7SLG
Ag in 7SLD
Ag in 7SDV
Ag in 7SLB