Zinc »
PDB 5c4j-5cdt »
5cby »
Zinc in PDB 5cby: ANCGR2 Dna Binding Domain - (+)Gre Complex
Protein crystallography data
The structure of ANCGR2 Dna Binding Domain - (+)Gre Complex, PDB code: 5cby
was solved by
W.H.Hudson,
E.A.Ortlund,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 36.74 / 2.00 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 131.644, 38.798, 98.250, 90.00, 119.65, 90.00 |
| R / Rfree (%) | 18.2 / 21.7 |
Zinc Binding Sites:
The binding sites of Zinc atom in the ANCGR2 Dna Binding Domain - (+)Gre Complex
(pdb code 5cby). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the ANCGR2 Dna Binding Domain - (+)Gre Complex, PDB code: 5cby:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the ANCGR2 Dna Binding Domain - (+)Gre Complex, PDB code: 5cby:
Jump to Zinc binding site number: 1; 2; 3; 4;
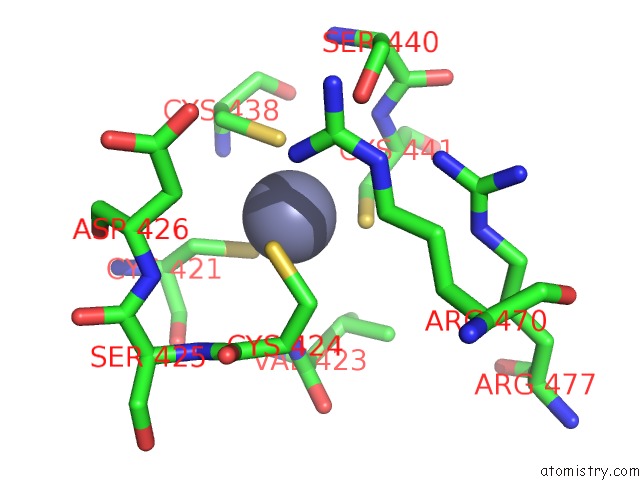
Zinc binding site 1 out of 4 in 5cby
Go back to
Zinc binding site 1 out
of 4 in the ANCGR2 Dna Binding Domain - (+)Gre Complex
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of ANCGR2 Dna Binding Domain - (+)Gre Complex within 5.0Å range:
|
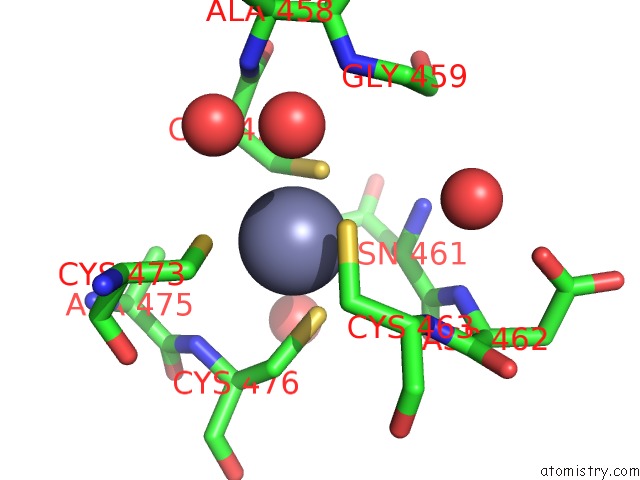
Zinc binding site 2 out of 4 in 5cby
Go back to
Zinc binding site 2 out
of 4 in the ANCGR2 Dna Binding Domain - (+)Gre Complex
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of ANCGR2 Dna Binding Domain - (+)Gre Complex within 5.0Å range:
|
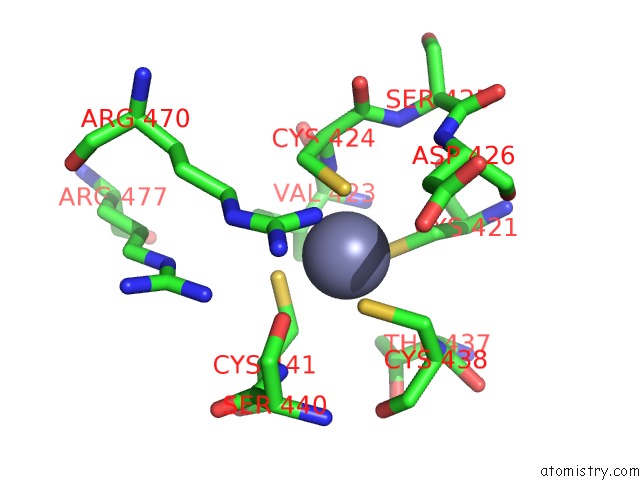
Zinc binding site 3 out of 4 in 5cby
Go back to
Zinc binding site 3 out
of 4 in the ANCGR2 Dna Binding Domain - (+)Gre Complex
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of ANCGR2 Dna Binding Domain - (+)Gre Complex within 5.0Å range:
|
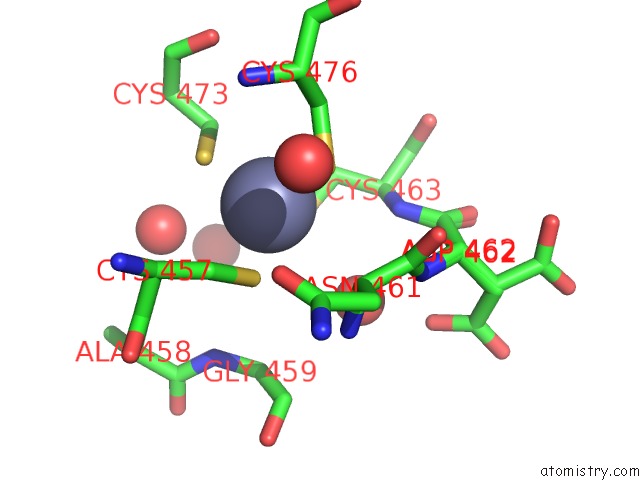
Zinc binding site 4 out of 4 in 5cby
Go back to
Zinc binding site 4 out
of 4 in the ANCGR2 Dna Binding Domain - (+)Gre Complex
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of ANCGR2 Dna Binding Domain - (+)Gre Complex within 5.0Å range:
|
Reference:
W.H.Hudson,
B.R.Kossmann,
I.M.De Vera,
S.W.Chuo,
E.R.Weikum,
G.N.Eick,
J.W.Thornton,
I.N.Ivanov,
D.J.Kojetin,
E.A.Ortlund.
Distal Substitutions Drive Divergent Dna Specificity Among Paralogous Transcription Factors Through Subdivision of Conformational Space. Proc.Natl.Acad.Sci.Usa V. 113 326 2016.
ISSN: ESSN 1091-6490
PubMed: 26715749
DOI: 10.1073/PNAS.1518960113
Page generated: Thu Aug 21 01:18:06 2025
ISSN: ESSN 1091-6490
PubMed: 26715749
DOI: 10.1073/PNAS.1518960113
Last articles
Zn in 5PLQZn in 5PLN
Zn in 5PLO
Zn in 5PLP
Zn in 5PLJ
Zn in 5PLL
Zn in 5PLK
Zn in 5PLM
Zn in 5PLH
Zn in 5PLI