Zinc »
PDB 4rvo-4tpj »
4s2t »
Zinc in PDB 4s2t: Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site
Protein crystallography data
The structure of Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site, PDB code: 4s2t
was solved by
S.Iyer,
P.La-Borde,
K.A.P.Payne,
M.R.Parsons,
A.J.Turner,
R.E.Isaac,
K.R.Acharya,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 38.52 / 2.15 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 140.590, 86.810, 113.130, 90.00, 115.96, 90.00 |
| R / Rfree (%) | 20.5 / 24.7 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site
(pdb code 4s2t). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 4 binding sites of Zinc where determined in the Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site, PDB code: 4s2t:
Jump to Zinc binding site number: 1; 2; 3; 4;
In total 4 binding sites of Zinc where determined in the Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site, PDB code: 4s2t:
Jump to Zinc binding site number: 1; 2; 3; 4;
Zinc binding site 1 out of 4 in 4s2t
Go back to
Zinc binding site 1 out
of 4 in the Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site
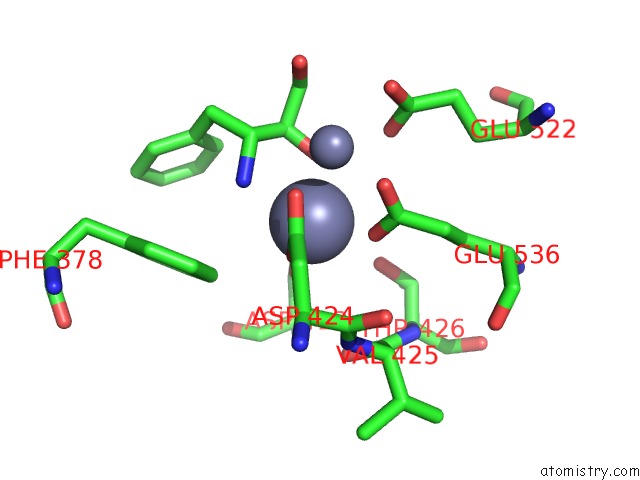
Mono view
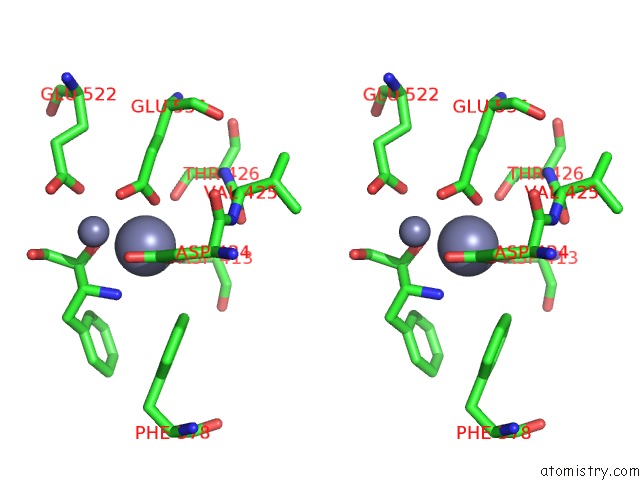
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site within 5.0Å range:
|
Zinc binding site 2 out of 4 in 4s2t
Go back to
Zinc binding site 2 out
of 4 in the Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site
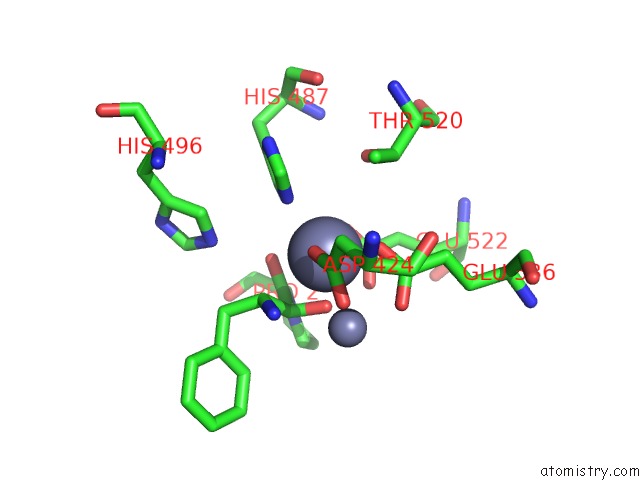
Mono view
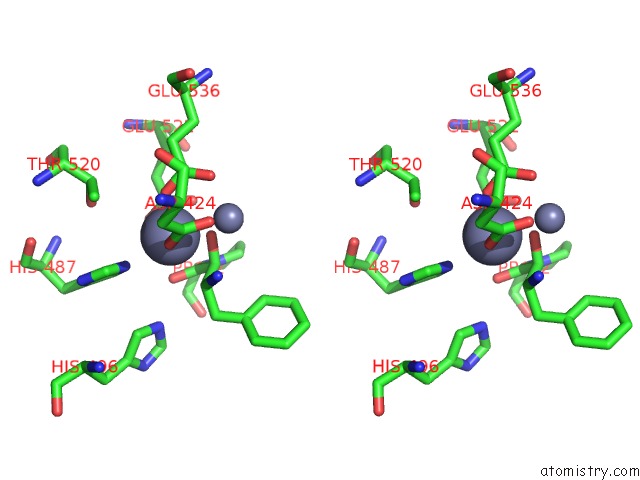
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site within 5.0Å range:
|
Zinc binding site 3 out of 4 in 4s2t
Go back to
Zinc binding site 3 out
of 4 in the Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site
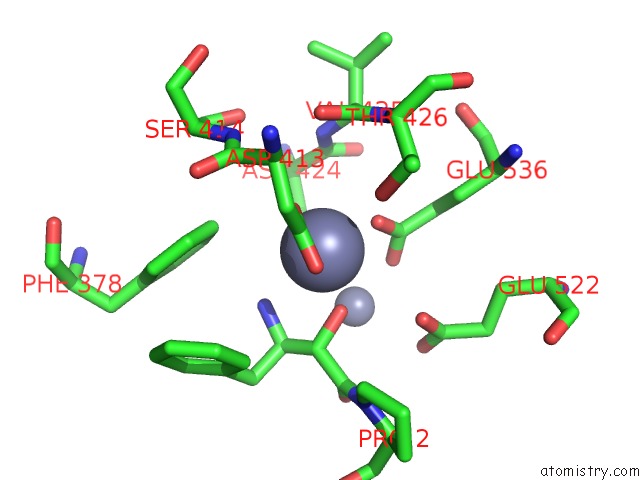
Mono view
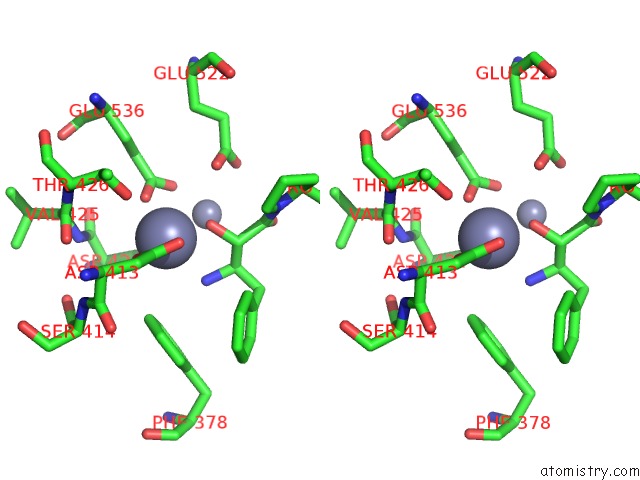
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site within 5.0Å range:
|
Zinc binding site 4 out of 4 in 4s2t
Go back to
Zinc binding site 4 out
of 4 in the Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site
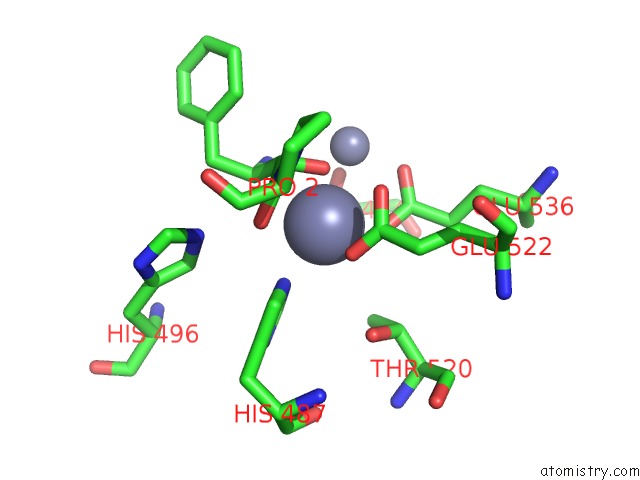
Mono view
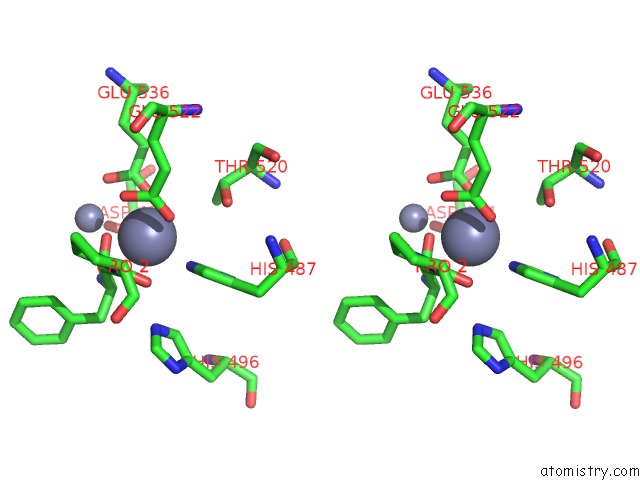
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site within 5.0Å range:
|
Reference:
S.Iyer,
P.J.La-Borde,
K.A.Payne,
M.R.Parsons,
A.J.Turner,
R.E.Isaac,
K.R.Acharya.
Crystal Structure of X-Prolyl Aminopeptidase From Caenorhabditis Elegans: A Cytosolic Enzyme with A Di-Nuclear Active Site. Febs Open Bio V. 5 292 2015.
ISSN: ESSN 2211-5463
PubMed: 25905034
DOI: 10.1016/J.FOB.2015.03.013
Page generated: Sun Oct 27 08:23:04 2024
ISSN: ESSN 2211-5463
PubMed: 25905034
DOI: 10.1016/J.FOB.2015.03.013
Last articles
As in 3FKGAs in 3FM4
As in 3FMU
As in 3ET6
As in 3ENZ
As in 3FJU
As in 3FCU
As in 3F7D
As in 3ERP
As in 3E7S