Zinc »
PDB 4i9z-4ijd »
4ibv »
Zinc in PDB 4ibv: Human P53 Core Domain with Hot Spot Mutation R273C and Second-Site Suppressor Mutation S240R in Sequence-Specific Complex with Dna
Protein crystallography data
The structure of Human P53 Core Domain with Hot Spot Mutation R273C and Second-Site Suppressor Mutation S240R in Sequence-Specific Complex with Dna, PDB code: 4ibv
was solved by
A.Eldar,
H.Rozenberg,
Y.Diskin-Posner,
Z.Shakked,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 47.23 / 2.10 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 137.631, 50.302, 34.008, 90.00, 93.46, 90.00 |
| R / Rfree (%) | 16.1 / 22.9 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Human P53 Core Domain with Hot Spot Mutation R273C and Second-Site Suppressor Mutation S240R in Sequence-Specific Complex with Dna
(pdb code 4ibv). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Human P53 Core Domain with Hot Spot Mutation R273C and Second-Site Suppressor Mutation S240R in Sequence-Specific Complex with Dna, PDB code: 4ibv:
In total only one binding site of Zinc was determined in the Human P53 Core Domain with Hot Spot Mutation R273C and Second-Site Suppressor Mutation S240R in Sequence-Specific Complex with Dna, PDB code: 4ibv:
Zinc binding site 1 out of 1 in 4ibv
Go back to
Zinc binding site 1 out
of 1 in the Human P53 Core Domain with Hot Spot Mutation R273C and Second-Site Suppressor Mutation S240R in Sequence-Specific Complex with Dna
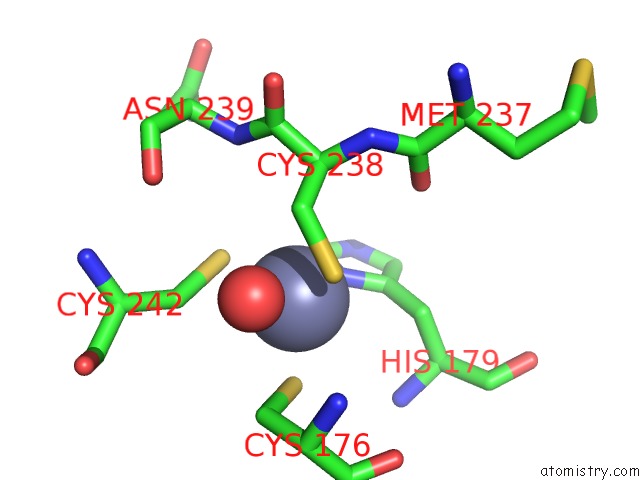
Mono view
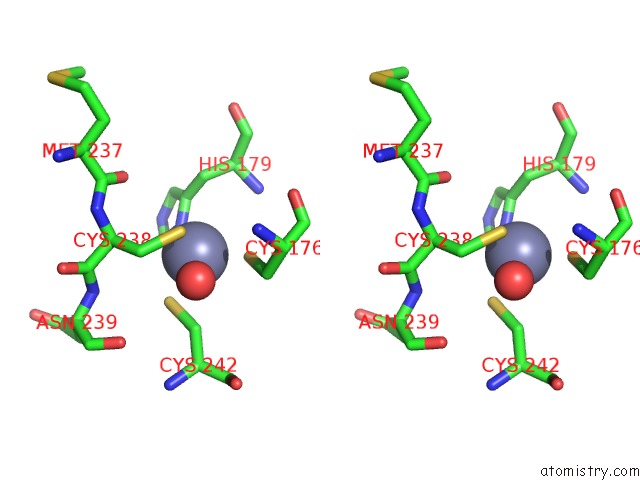
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Human P53 Core Domain with Hot Spot Mutation R273C and Second-Site Suppressor Mutation S240R in Sequence-Specific Complex with Dna within 5.0Å range:
|
Reference:
A.Eldar,
H.Rozenberg,
Y.Diskin-Posner,
R.Rohs,
Z.Shakked.
Structural Studies of P53 Inactivation By Dna-Contact Mutations and Its Rescue By Suppressor Mutations Via Alternative Protein-Dna Interactions. Nucleic Acids Res. V. 41 8748 2013.
ISSN: ISSN 0305-1048
PubMed: 23863845
DOI: 10.1093/NAR/GKT630
Page generated: Wed Aug 20 18:49:27 2025
ISSN: ISSN 0305-1048
PubMed: 23863845
DOI: 10.1093/NAR/GKT630
Last articles
Zn in 5A7MZn in 5ABA
Zn in 5AB2
Zn in 5AB9
Zn in 5AB0
Zn in 5AAZ
Zn in 5AAS
Zn in 5AAY
Zn in 5A87
Zn in 5AAQ