Zinc »
PDB 4cis-4csp »
4cn7 »
Zinc in PDB 4cn7: Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element
Protein crystallography data
The structure of Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element, PDB code: 4cn7
was solved by
A.G.Mcewen,
P.Poussin-Courmontagne,
J.Osz,
N.Rochel,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.82 / 2.34 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 37.630, 65.350, 209.120, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.31 / 20.93 |
Other elements in 4cn7:
The structure of Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element also contains other interesting chemical elements:
| Chlorine | (Cl) | 3 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element
(pdb code 4cn7). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 8 binding sites of Zinc where determined in the Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element, PDB code: 4cn7:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Zinc where determined in the Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element, PDB code: 4cn7:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Zinc binding site 1 out of 8 in 4cn7
Go back to
Zinc binding site 1 out
of 8 in the Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element
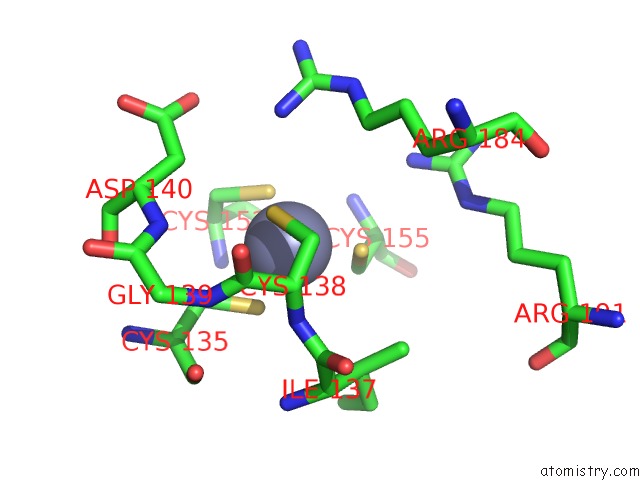
Mono view
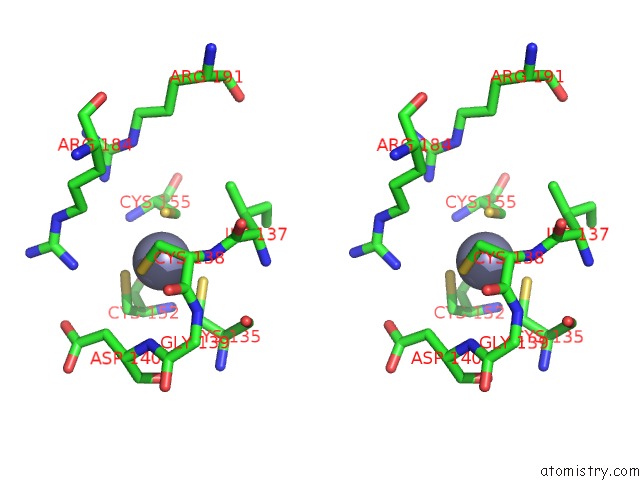
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element within 5.0Å range:
|
Zinc binding site 2 out of 8 in 4cn7
Go back to
Zinc binding site 2 out
of 8 in the Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element

Mono view
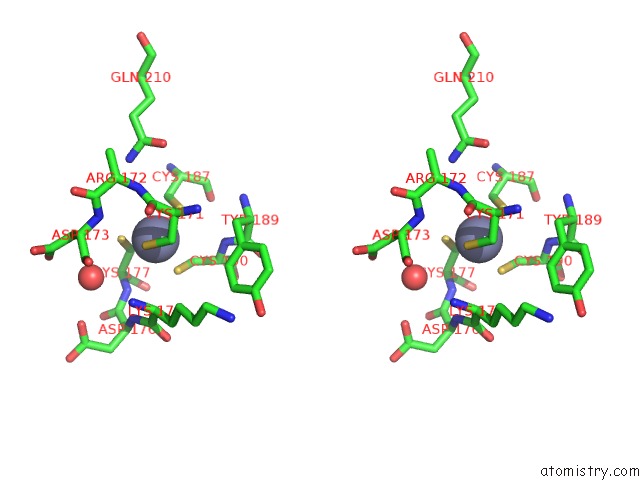
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element within 5.0Å range:
|
Zinc binding site 3 out of 8 in 4cn7
Go back to
Zinc binding site 3 out
of 8 in the Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element
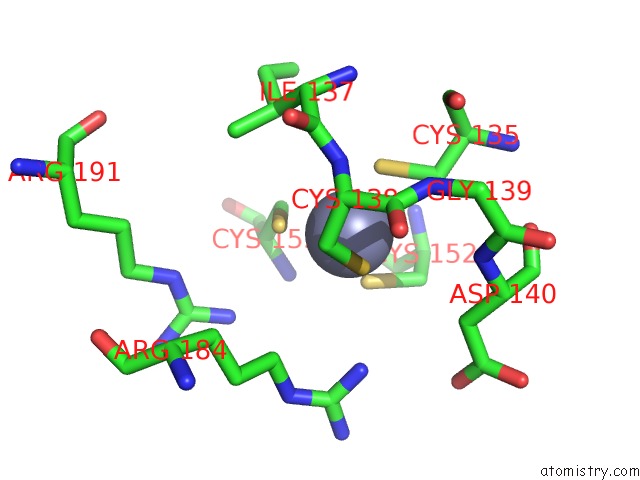
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element within 5.0Å range:
|
Zinc binding site 4 out of 8 in 4cn7
Go back to
Zinc binding site 4 out
of 8 in the Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element
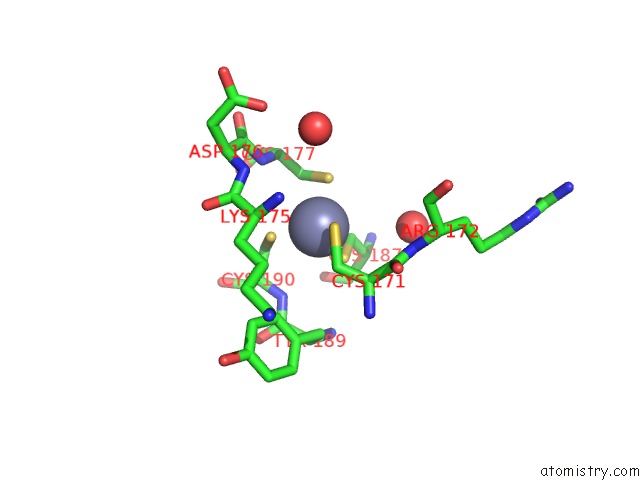
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element within 5.0Å range:
|
Zinc binding site 5 out of 8 in 4cn7
Go back to
Zinc binding site 5 out
of 8 in the Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element within 5.0Å range:
|
Zinc binding site 6 out of 8 in 4cn7
Go back to
Zinc binding site 6 out
of 8 in the Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element within 5.0Å range:
|
Zinc binding site 7 out of 8 in 4cn7
Go back to
Zinc binding site 7 out
of 8 in the Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element within 5.0Å range:
|
Zinc binding site 8 out of 8 in 4cn7
Go back to
Zinc binding site 8 out
of 8 in the Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element

Mono view
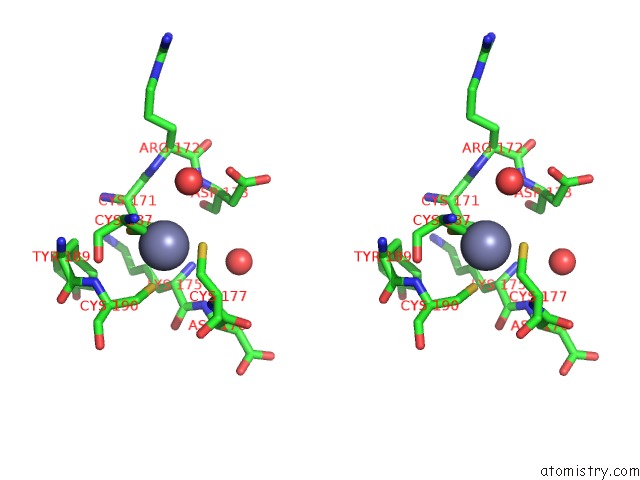
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 8 of Crystal Structure of the Human Retinoid X Receptor Dna- Binding Domain Bound to An Idealized DR1 Response Element within 5.0Å range:
|
Reference:
J.Osz,
A.G.Mcewen,
P.Poussin-Courmontagne,
E.Moutier,
C.Birck,
I.Davidson,
D.Moras,
N.Rochel.
Structural Basis of Natural Promoter Recognition By the Retinoid X Nuclear Receptor. Sci.Rep. V. 5 8216 2015.
ISSN: ISSN 2045-2322
PubMed: 25645674
DOI: 10.1038/SREP08216
Page generated: Sat Oct 26 20:49:34 2024
ISSN: ISSN 2045-2322
PubMed: 25645674
DOI: 10.1038/SREP08216
Last articles
As in 1IHUAs in 1I83
As in 1I9T
As in 1I9S
As in 1HYV
As in 1I19
As in 1GLJ
As in 1GQ8
As in 1GR0
As in 1FU1