Zinc »
PDB 3qmc-3qzv »
3qz2 »
Zinc in PDB 3qz2: The Structure of Cysteine-Free Human Insulin Degrading Enzyme
Enzymatic activity of The Structure of Cysteine-Free Human Insulin Degrading Enzyme
All present enzymatic activity of The Structure of Cysteine-Free Human Insulin Degrading Enzyme:
3.4.24.56;
3.4.24.56;
Protein crystallography data
The structure of The Structure of Cysteine-Free Human Insulin Degrading Enzyme, PDB code: 3qz2
was solved by
Q.Guo,
W.J.Tang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 3.20 |
| Space group | P 65 |
| Cell size a, b, c (Å), α, β, γ (°) | 262.073, 262.073, 90.838, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 17.2 / 24.1 |
Zinc Binding Sites:
The binding sites of Zinc atom in the The Structure of Cysteine-Free Human Insulin Degrading Enzyme
(pdb code 3qz2). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the The Structure of Cysteine-Free Human Insulin Degrading Enzyme, PDB code: 3qz2:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the The Structure of Cysteine-Free Human Insulin Degrading Enzyme, PDB code: 3qz2:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 3qz2
Go back to
Zinc binding site 1 out
of 2 in the The Structure of Cysteine-Free Human Insulin Degrading Enzyme
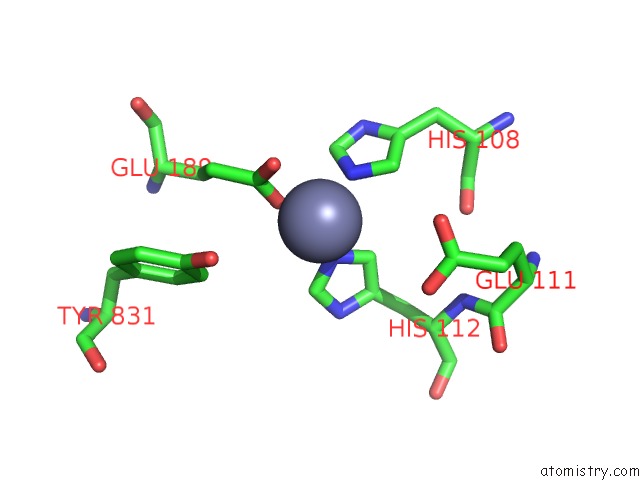
Mono view
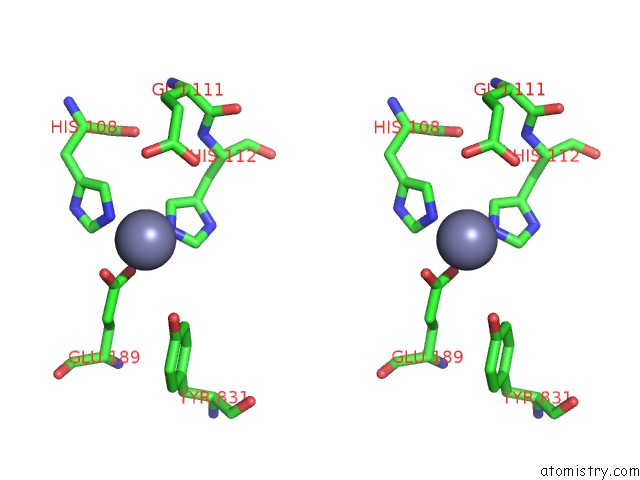
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of The Structure of Cysteine-Free Human Insulin Degrading Enzyme within 5.0Å range:
|
Zinc binding site 2 out of 2 in 3qz2
Go back to
Zinc binding site 2 out
of 2 in the The Structure of Cysteine-Free Human Insulin Degrading Enzyme
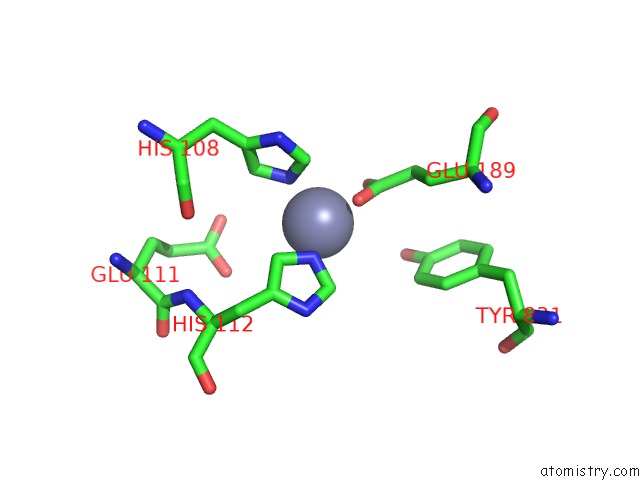
Mono view
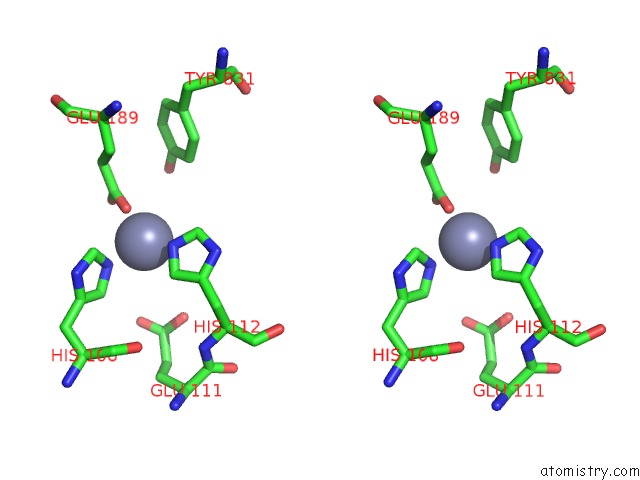
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of The Structure of Cysteine-Free Human Insulin Degrading Enzyme within 5.0Å range:
|
Reference:
J.Charton,
M.Gauriot,
Q.Guo,
N.Hennuyer,
X.Marechal,
J.Dumont,
M.Hamdane,
V.Pottiez,
V.Landry,
O.Sperandio,
M.Flipo,
L.Buee,
B.Staels,
F.Leroux,
W.J.Tang,
B.Deprez,
R.Deprez-Poulain.
Imidazole-Derived 2-[N-Carbamoylmethyl-Alkylamino]Acetic Acids, Substrate-Dependent Modulators of Insulin-Degrading Enzyme in Amyloid-Beta Hydrolysis. Eur.J.Med.Chem. V. 79 184 2014.
ISSN: ISSN 0223-5234
PubMed: 24735644
DOI: 10.1016/J.EJMECH.2014.04.009
Page generated: Sat Oct 26 12:26:49 2024
ISSN: ISSN 0223-5234
PubMed: 24735644
DOI: 10.1016/J.EJMECH.2014.04.009
Last articles
As in 1ZZSAs in 1ZZT
As in 1ZV8
As in 1Y0X
As in 1YHC
As in 1XZX
As in 1Y9A
As in 1WN5
As in 1Y0R
As in 1XSL