Zinc »
PDB 3qmd-3r0d »
3qwv »
Zinc in PDB 3qwv: Crystal Structure of Histone Lysine Methyltransferase SMYD2 in Complex with the Cofactor Product Adohcy
Protein crystallography data
The structure of Crystal Structure of Histone Lysine Methyltransferase SMYD2 in Complex with the Cofactor Product Adohcy, PDB code: 3qwv
was solved by
Y.Jiang,
N.Sirinupong,
J.Brunzelle,
Z.Yang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 45.24 / 2.03 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 57.871, 75.043, 113.409, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 17.3 / 21.5 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Histone Lysine Methyltransferase SMYD2 in Complex with the Cofactor Product Adohcy
(pdb code 3qwv). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 3 binding sites of Zinc where determined in the Crystal Structure of Histone Lysine Methyltransferase SMYD2 in Complex with the Cofactor Product Adohcy, PDB code: 3qwv:
Jump to Zinc binding site number: 1; 2; 3;
In total 3 binding sites of Zinc where determined in the Crystal Structure of Histone Lysine Methyltransferase SMYD2 in Complex with the Cofactor Product Adohcy, PDB code: 3qwv:
Jump to Zinc binding site number: 1; 2; 3;
Zinc binding site 1 out of 3 in 3qwv
Go back to
Zinc binding site 1 out
of 3 in the Crystal Structure of Histone Lysine Methyltransferase SMYD2 in Complex with the Cofactor Product Adohcy
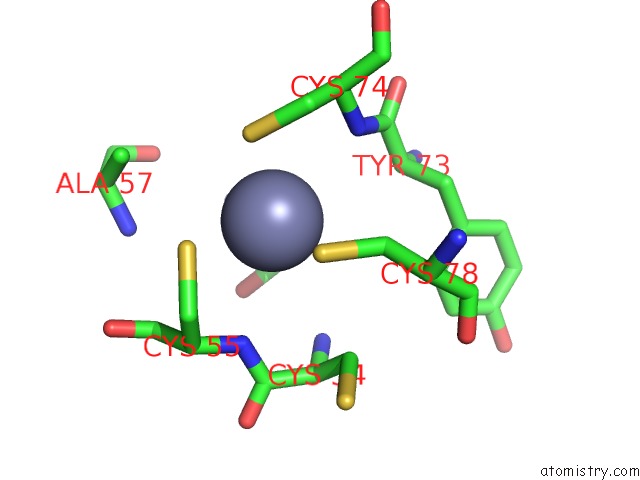
Mono view
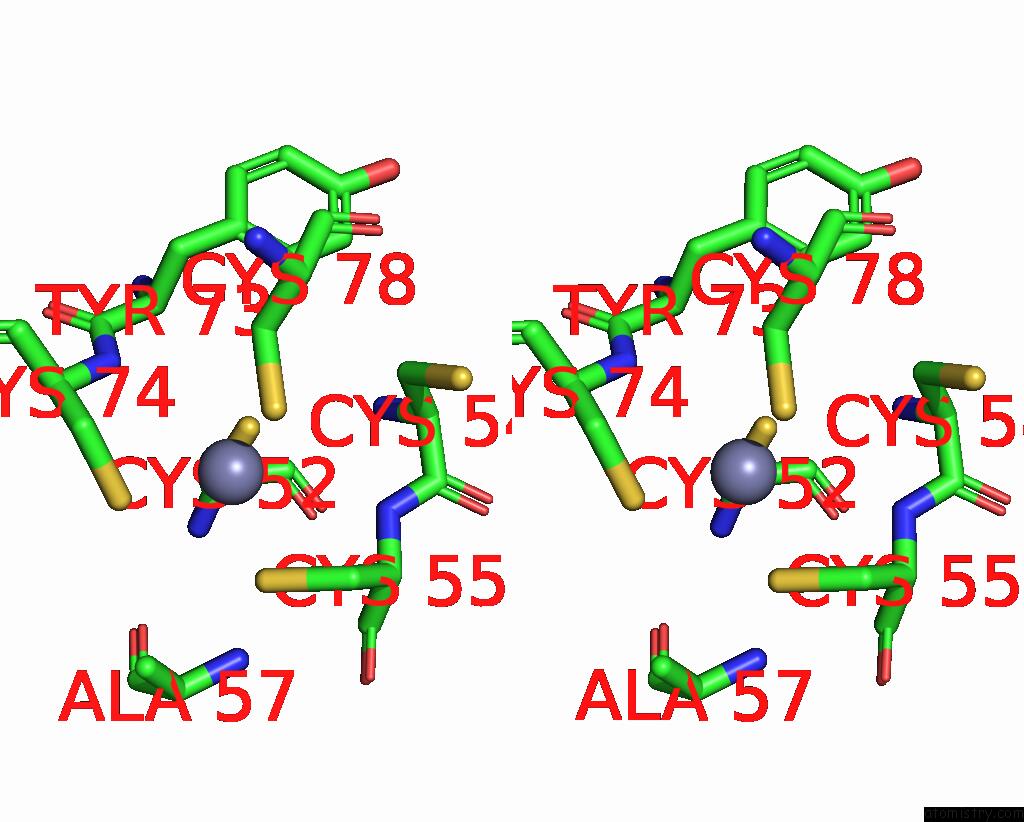
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Histone Lysine Methyltransferase SMYD2 in Complex with the Cofactor Product Adohcy within 5.0Å range:
|
Zinc binding site 2 out of 3 in 3qwv
Go back to
Zinc binding site 2 out
of 3 in the Crystal Structure of Histone Lysine Methyltransferase SMYD2 in Complex with the Cofactor Product Adohcy
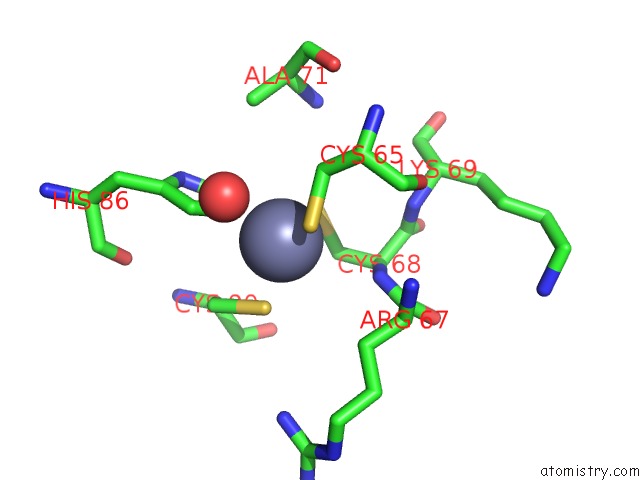
Mono view
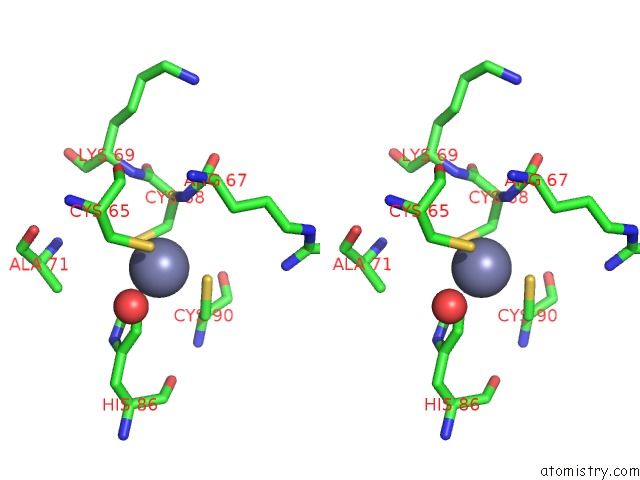
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Histone Lysine Methyltransferase SMYD2 in Complex with the Cofactor Product Adohcy within 5.0Å range:
|
Zinc binding site 3 out of 3 in 3qwv
Go back to
Zinc binding site 3 out
of 3 in the Crystal Structure of Histone Lysine Methyltransferase SMYD2 in Complex with the Cofactor Product Adohcy
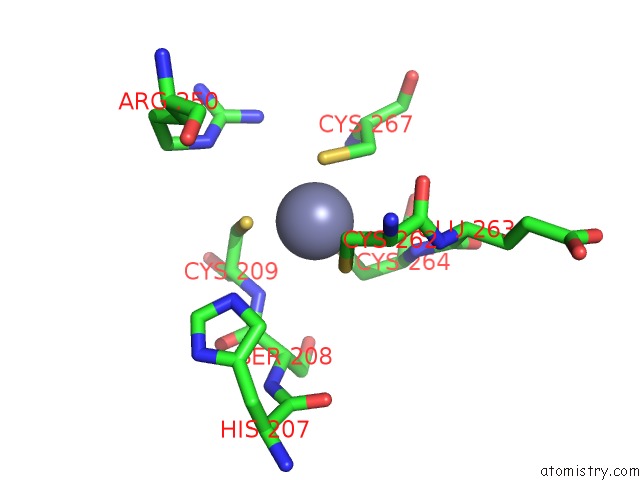
Mono view
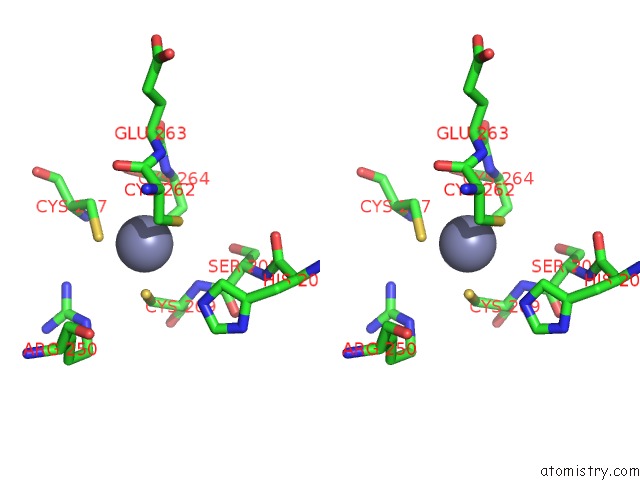
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of Histone Lysine Methyltransferase SMYD2 in Complex with the Cofactor Product Adohcy within 5.0Å range:
|
Reference:
Y.Jiang,
N.Sirinupong,
J.Brunzelle,
Z.Yang.
Crystal Structures of Histone and P53 Methyltransferase SMYD2 Reveal A Conformational Flexibility of the Autoinhibitory C-Terminal Domain. Plos One V. 6 21640 2011.
ISSN: ESSN 1932-6203
PubMed: 21738746
DOI: 10.1371/JOURNAL.PONE.0021640
Page generated: Wed Aug 20 13:23:26 2025
ISSN: ESSN 1932-6203
PubMed: 21738746
DOI: 10.1371/JOURNAL.PONE.0021640
Last articles
Zn in 4ISAZn in 4IS9
Zn in 4IOU
Zn in 4IR0
Zn in 4IPP
Zn in 4IN9
Zn in 4IOF
Zn in 4INS
Zn in 4IMX
Zn in 4IMU