Zinc »
PDB 3bof-3c52 »
3bxq »
Zinc in PDB 3bxq: The Structure of A Mutant Insulin Uncouples Receptor Binding From Protein Allostery. An Electrostatic Block to the Tr Transition
Protein crystallography data
The structure of The Structure of A Mutant Insulin Uncouples Receptor Binding From Protein Allostery. An Electrostatic Block to the Tr Transition, PDB code: 3bxq
was solved by
Z.L.Wan,
K.Huang,
S.Q.Hu,
J.Whittaker,
M.A.Weiss,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.62 / 1.30 |
| Space group | H 3 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.390, 81.390, 34.000, 90.00, 90.00, 120.00 |
| R / Rfree (%) | 20.6 / 23.2 |
Zinc Binding Sites:
The binding sites of Zinc atom in the The Structure of A Mutant Insulin Uncouples Receptor Binding From Protein Allostery. An Electrostatic Block to the Tr Transition
(pdb code 3bxq). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the The Structure of A Mutant Insulin Uncouples Receptor Binding From Protein Allostery. An Electrostatic Block to the Tr Transition, PDB code: 3bxq:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the The Structure of A Mutant Insulin Uncouples Receptor Binding From Protein Allostery. An Electrostatic Block to the Tr Transition, PDB code: 3bxq:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 3bxq
Go back to
Zinc binding site 1 out
of 2 in the The Structure of A Mutant Insulin Uncouples Receptor Binding From Protein Allostery. An Electrostatic Block to the Tr Transition
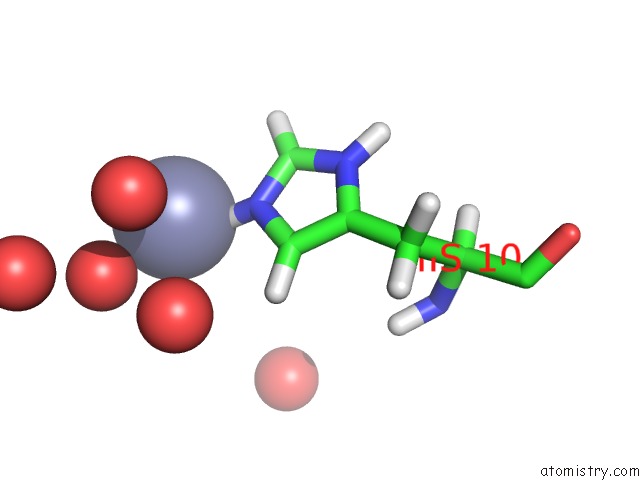
Mono view
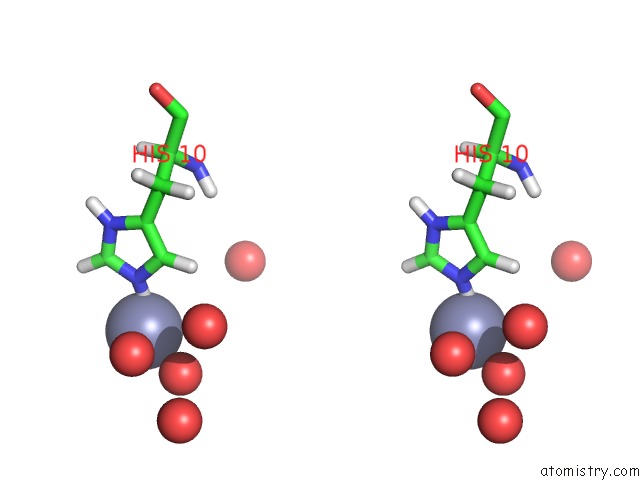
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of The Structure of A Mutant Insulin Uncouples Receptor Binding From Protein Allostery. An Electrostatic Block to the Tr Transition within 5.0Å range:
|
Zinc binding site 2 out of 2 in 3bxq
Go back to
Zinc binding site 2 out
of 2 in the The Structure of A Mutant Insulin Uncouples Receptor Binding From Protein Allostery. An Electrostatic Block to the Tr Transition
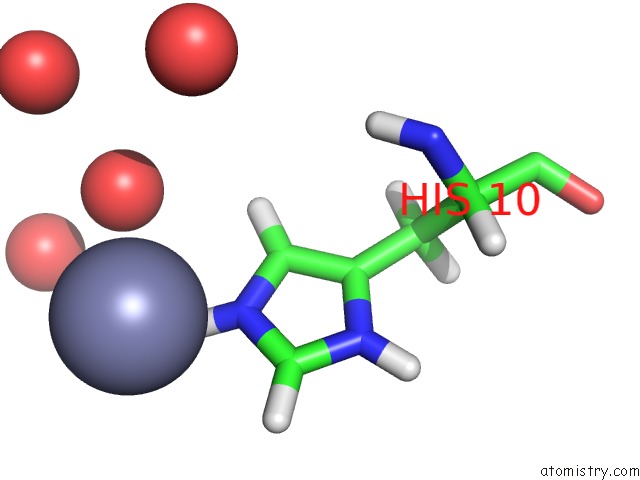
Mono view
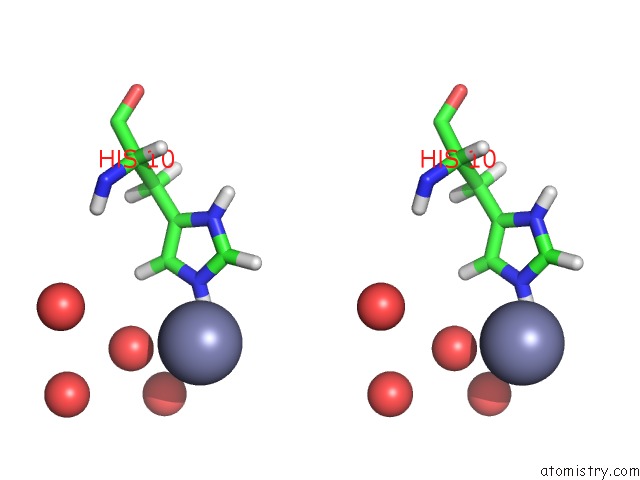
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of The Structure of A Mutant Insulin Uncouples Receptor Binding From Protein Allostery. An Electrostatic Block to the Tr Transition within 5.0Å range:
|
Reference:
Q.X.Hua,
S.H.Nakagawa,
W.Jia,
K.Huang,
N.B.Phillips,
S.Q.Hu,
M.A.Weiss.
Design of An Active Ultrastable Single-Chain Insulin Analog: Synthesis, Structure, and Therapeutic Implications. J.Biol.Chem. V. 283 14703 2008.
ISSN: ISSN 0021-9258
PubMed: 18332129
DOI: 10.1074/JBC.M800313200
Page generated: Wed Aug 20 08:05:51 2025
ISSN: ISSN 0021-9258
PubMed: 18332129
DOI: 10.1074/JBC.M800313200
Last articles
Zn in 3M97Zn in 3M79
Zn in 3M98
Zn in 3M7P
Zn in 3M85
Zn in 3M96
Zn in 3M7N
Zn in 3M7K
Zn in 3M6R
Zn in 3M6O