Zinc »
PDB 2xy9-2y7g »
2y6i »
Zinc in PDB 2y6i: Crystal Structure of Collagenase G From Clostridium Histolyticum in Complex with Isoamylphosphonyl-Gly-Pro-Ala at 3.25 Angstrom Resolution
Enzymatic activity of Crystal Structure of Collagenase G From Clostridium Histolyticum in Complex with Isoamylphosphonyl-Gly-Pro-Ala at 3.25 Angstrom Resolution
All present enzymatic activity of Crystal Structure of Collagenase G From Clostridium Histolyticum in Complex with Isoamylphosphonyl-Gly-Pro-Ala at 3.25 Angstrom Resolution:
3.4.24.3;
3.4.24.3;
Protein crystallography data
The structure of Crystal Structure of Collagenase G From Clostridium Histolyticum in Complex with Isoamylphosphonyl-Gly-Pro-Ala at 3.25 Angstrom Resolution, PDB code: 2y6i
was solved by
U.Eckhard,
H.Brandstetter,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 93.28 / 3.25 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 58.120, 108.840, 181.030, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.64 / 26.633 |
Zinc Binding Sites:
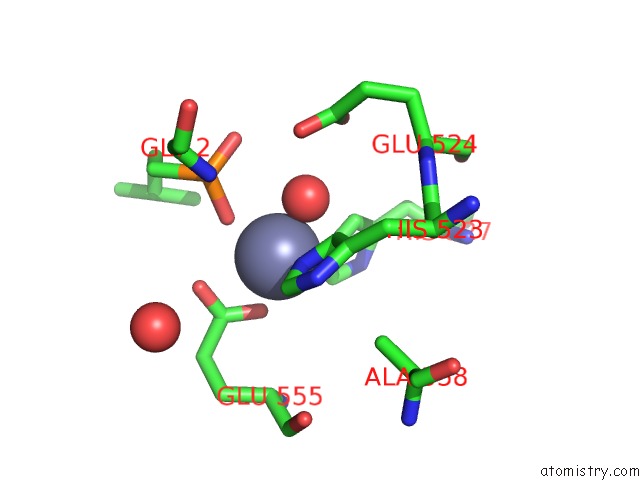
The binding sites of Zinc atom in the Crystal Structure of Collagenase G From Clostridium Histolyticum in Complex with Isoamylphosphonyl-Gly-Pro-Ala at 3.25 Angstrom Resolution
(pdb code 2y6i). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Crystal Structure of Collagenase G From Clostridium Histolyticum in Complex with Isoamylphosphonyl-Gly-Pro-Ala at 3.25 Angstrom Resolution, PDB code: 2y6i:
In total only one binding site of Zinc was determined in the Crystal Structure of Collagenase G From Clostridium Histolyticum in Complex with Isoamylphosphonyl-Gly-Pro-Ala at 3.25 Angstrom Resolution, PDB code: 2y6i:
Zinc binding site 1 out of 1 in 2y6i
Go back to
Zinc binding site 1 out
of 1 in the Crystal Structure of Collagenase G From Clostridium Histolyticum in Complex with Isoamylphosphonyl-Gly-Pro-Ala at 3.25 Angstrom Resolution
Mono view
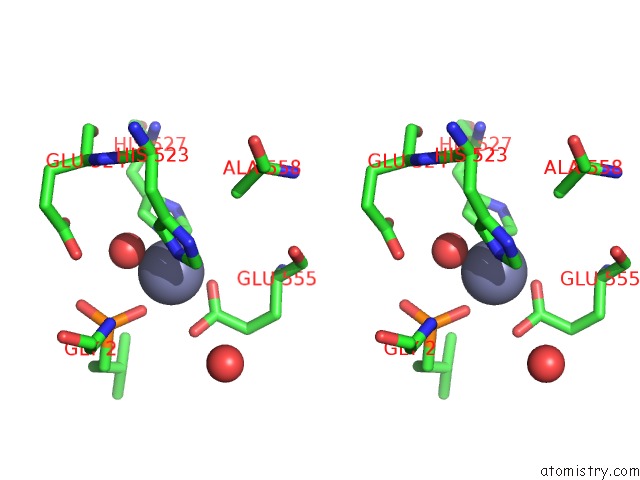
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Collagenase G From Clostridium Histolyticum in Complex with Isoamylphosphonyl-Gly-Pro-Ala at 3.25 Angstrom Resolution within 5.0Å range:
|
Reference:
U.Eckhard,
E.Schoenauer,
D.Nuess,
H.Brandstetter.
Structure of Collagenase G Reveals A Chew-and -Digest Mechanism of Bacterial Collagenolysis Nat.Struct.Mol.Biol. V. 18 1109 2011.
ISSN: ISSN 1545-9993
PubMed: 21947205
DOI: 10.1038/NSMB.2127
Page generated: Wed Aug 20 07:02:27 2025
ISSN: ISSN 1545-9993
PubMed: 21947205
DOI: 10.1038/NSMB.2127
Last articles
Zn in 3QN9Zn in 3QMG
Zn in 3QMH
Zn in 3QMI
Zn in 3QMD
Zn in 3QM3
Zn in 3QLN
Zn in 3QMC
Zn in 3QMB
Zn in 3QLC