Zinc »
PDB 2xl9-2xy4 »
2xs2 »
Zinc in PDB 2xs2: Crystal Structure of the Rrm Domain of Mouse Deleted in Azoospermia- Like in Complex with Rna, Uuguucuu
Protein crystallography data
The structure of Crystal Structure of the Rrm Domain of Mouse Deleted in Azoospermia- Like in Complex with Rna, Uuguucuu, PDB code: 2xs2
was solved by
H.T.Jenkins,
T.A.Edwards,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 27.41 / 1.35 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 38.100, 52.060, 60.490, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 16.1 / 19.3 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of the Rrm Domain of Mouse Deleted in Azoospermia- Like in Complex with Rna, Uuguucuu
(pdb code 2xs2). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Crystal Structure of the Rrm Domain of Mouse Deleted in Azoospermia- Like in Complex with Rna, Uuguucuu, PDB code: 2xs2:
In total only one binding site of Zinc was determined in the Crystal Structure of the Rrm Domain of Mouse Deleted in Azoospermia- Like in Complex with Rna, Uuguucuu, PDB code: 2xs2:
Zinc binding site 1 out of 1 in 2xs2
Go back to
Zinc binding site 1 out
of 1 in the Crystal Structure of the Rrm Domain of Mouse Deleted in Azoospermia- Like in Complex with Rna, Uuguucuu
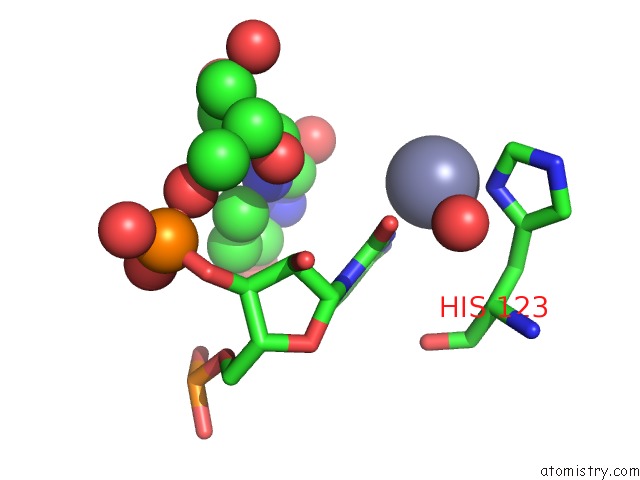
Mono view
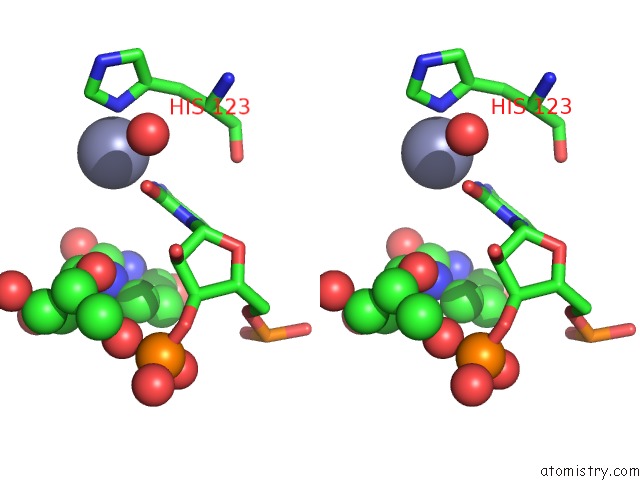
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of the Rrm Domain of Mouse Deleted in Azoospermia- Like in Complex with Rna, Uuguucuu within 5.0Å range:
|
Reference:
H.T.Jenkins,
B.Malkova,
T.A.Edwards.
Kinked Beta-Strands Mediate High-Affinity Recognition of Mrna Targets By the Germ-Cell Regulator Dazl. Proc. Natl. Acad. Sci. V. 108 18266 2011U.S.A..
ISSN: ESSN 1091-6490
PubMed: 22021443
DOI: 10.1073/PNAS.1105211108
Page generated: Wed Aug 20 06:52:02 2025
ISSN: ESSN 1091-6490
PubMed: 22021443
DOI: 10.1073/PNAS.1105211108
Last articles
Zn in 3HXEZn in 3HXD
Zn in 3HXC
Zn in 3HWP
Zn in 3HW7
Zn in 3HXB
Zn in 3HUV
Zn in 3HUD
Zn in 3HSV
Zn in 3HTR