Zinc »
PDB 2q42-2qmu »
2qii »
Zinc in PDB 2qii: Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0
Enzymatic activity of Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0
All present enzymatic activity of Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0:
2.4.2.29;
2.4.2.29;
Protein crystallography data
The structure of Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0, PDB code: 2qii
was solved by
N.Tidten,
R.Brenk,
A.Heine,
K.Reuter,
G.Klebe,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20.00 / 1.70 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 90.622, 65.535, 70.325, 90.00, 96.47, 90.00 |
| R / Rfree (%) | 16.6 / 21.3 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0
(pdb code 2qii). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0, PDB code: 2qii:
In total only one binding site of Zinc was determined in the Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0, PDB code: 2qii:
Zinc binding site 1 out of 1 in 2qii
Go back to
Zinc binding site 1 out
of 1 in the Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0
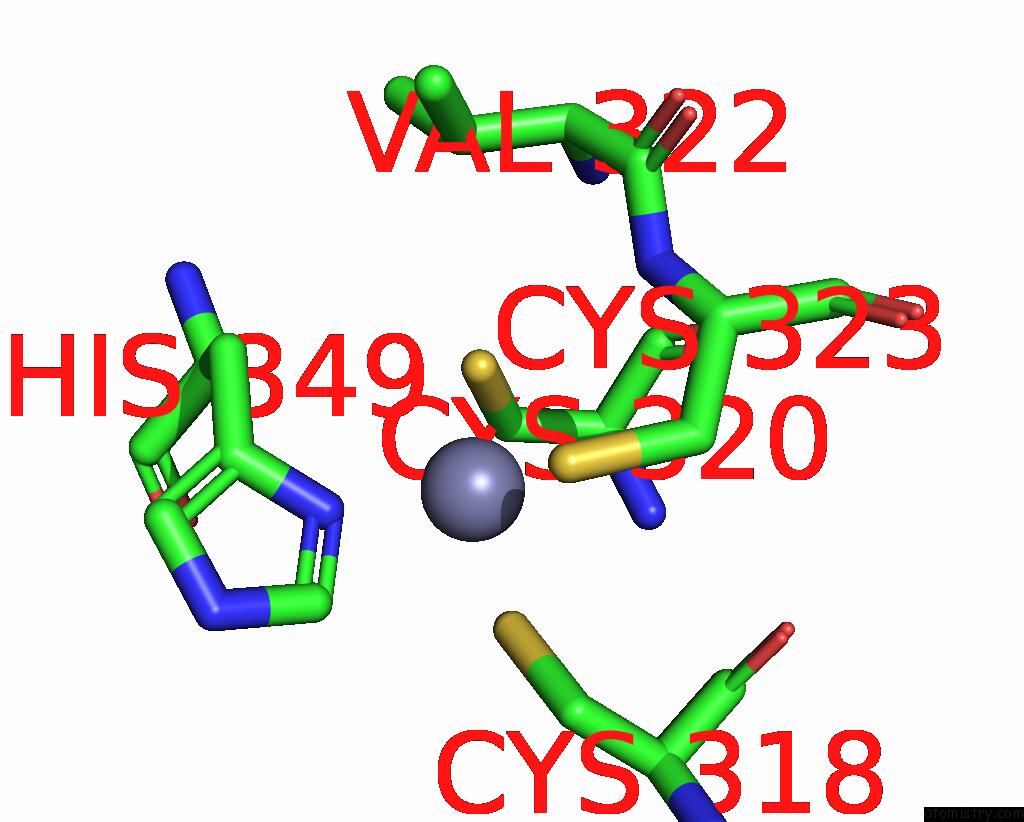
Mono view
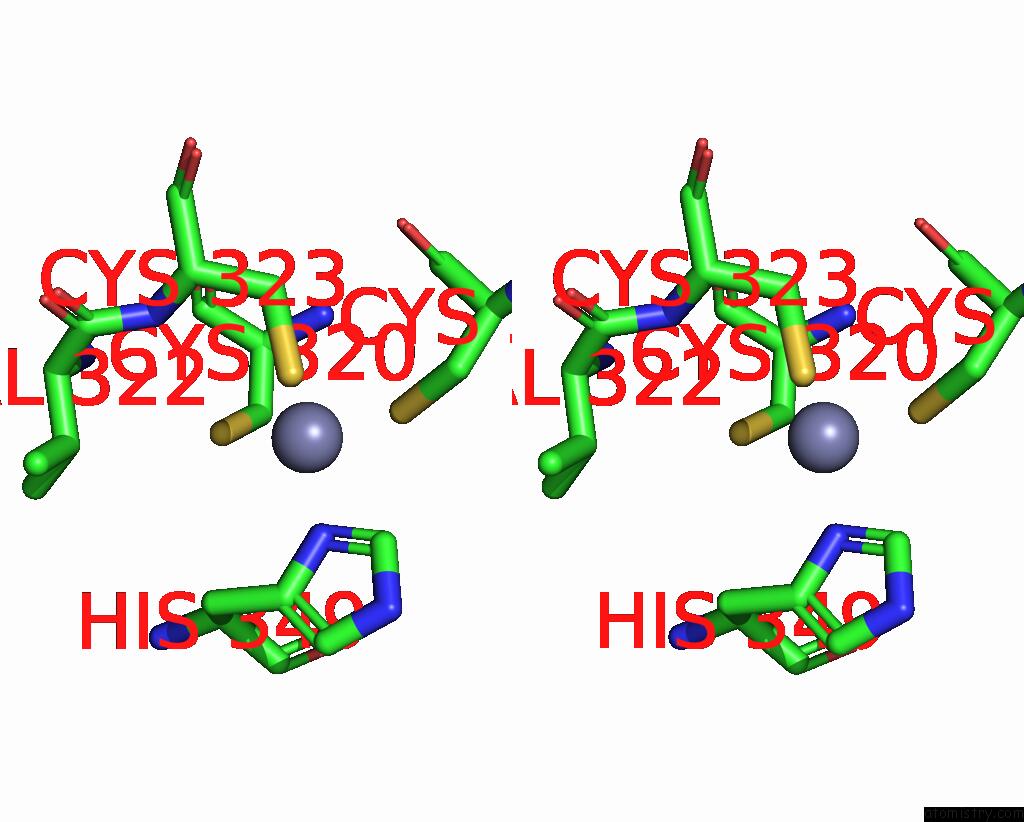
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Trna-Guanine Transglycosylase (Tgt) From Zymomonas Mobilis Complexed with Archaeosine Precursor, PREQ0 within 5.0Å range:
|
Reference:
N.Tidten,
B.Stengl,
A.Heine,
G.A.Garcia,
G.Klebe,
K.Reuter.
Glutamate Versus Glutamine Exchange Swaps Substrate Selectivity in Trna-Guanine Transglycosylase: Insight Into the Regulation of Substrate Selectivity By Kinetic and Crystallographic Studies. J.Mol.Biol. V. 374 764 2007.
ISSN: ISSN 0022-2836
PubMed: 17949745
DOI: 10.1016/J.JMB.2007.09.062
Page generated: Wed Aug 20 05:24:26 2025
ISSN: ISSN 0022-2836
PubMed: 17949745
DOI: 10.1016/J.JMB.2007.09.062
Last articles
Zn in 3GLRZn in 3GLJ
Zn in 3GL6
Zn in 3GJ9
Zn in 3GKL
Zn in 3GKY
Zn in 3GJ7
Zn in 3GJ8
Zn in 3GJN
Zn in 3GJ4