Zinc »
PDB 2q42-2qmu »
2qg9 »
Zinc in PDB 2qg9: Structure of A Regulatory Subunit Mutant D19A of Atcase From E. Coli
Enzymatic activity of Structure of A Regulatory Subunit Mutant D19A of Atcase From E. Coli
All present enzymatic activity of Structure of A Regulatory Subunit Mutant D19A of Atcase From E. Coli:
2.1.3.2;
2.1.3.2;
Protein crystallography data
The structure of Structure of A Regulatory Subunit Mutant D19A of Atcase From E. Coli, PDB code: 2qg9
was solved by
B.Stec,
M.K.Williams,
K.A.Stieglitz,
E.R.Kantrowitz,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 35.00 / 2.70 |
| Space group | P 3 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 122.290, 122.290, 142.410, 90.00, 90.00, 120.00 |
| R / Rfree (%) | n/a / 24.4 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Structure of A Regulatory Subunit Mutant D19A of Atcase From E. Coli
(pdb code 2qg9). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Structure of A Regulatory Subunit Mutant D19A of Atcase From E. Coli, PDB code: 2qg9:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Structure of A Regulatory Subunit Mutant D19A of Atcase From E. Coli, PDB code: 2qg9:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 2qg9
Go back to
Zinc binding site 1 out
of 2 in the Structure of A Regulatory Subunit Mutant D19A of Atcase From E. Coli
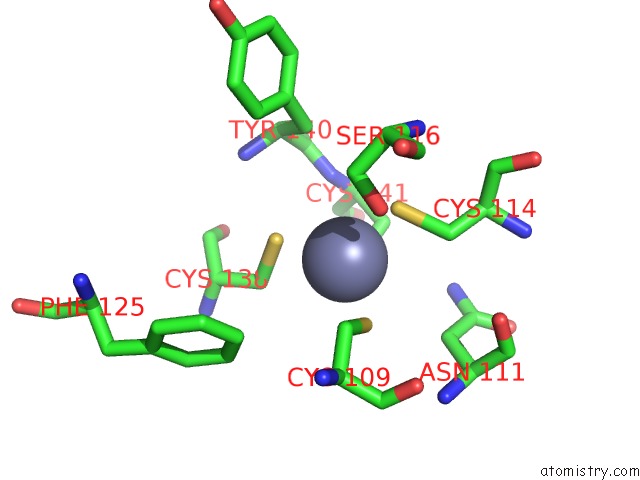
Mono view
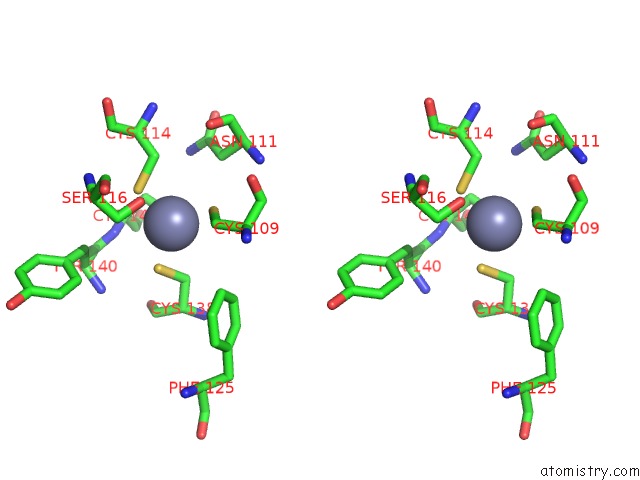
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structure of A Regulatory Subunit Mutant D19A of Atcase From E. Coli within 5.0Å range:
|
Zinc binding site 2 out of 2 in 2qg9
Go back to
Zinc binding site 2 out
of 2 in the Structure of A Regulatory Subunit Mutant D19A of Atcase From E. Coli
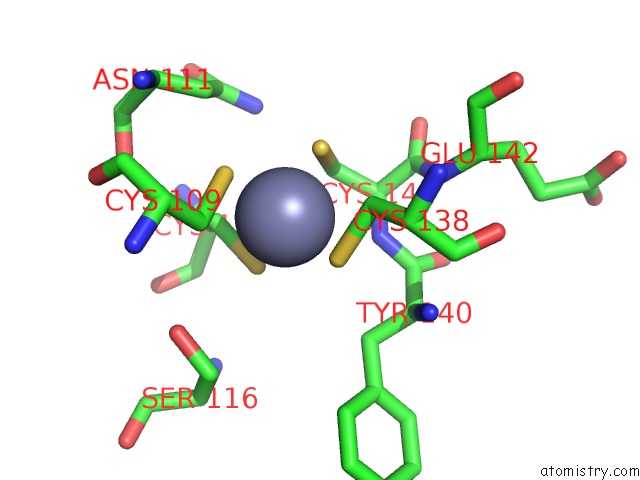
Mono view
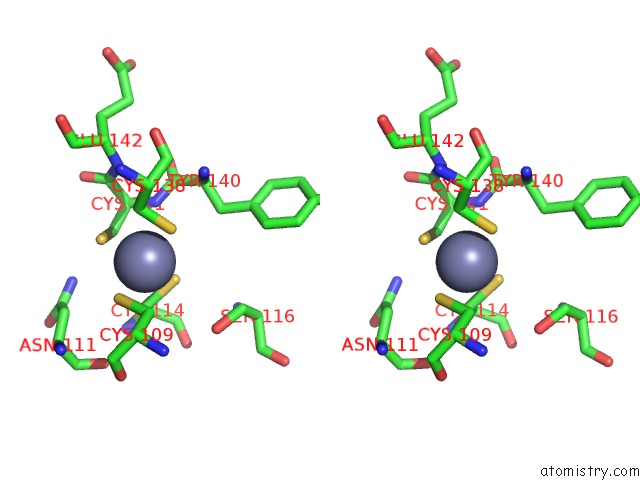
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structure of A Regulatory Subunit Mutant D19A of Atcase From E. Coli within 5.0Å range:
|
Reference:
B.Stec,
M.K.Williams,
K.A.Stieglitz,
E.R.Kantrowitz.
Comparison of Two T-State Structures of Regulatory-Chain Mutants of Escherichia Coli Aspartate Transcarbamoylase Suggests That HIS20 and ASP19 Modulate the Response to Heterotropic Effectors. Acta Crystallogr.,Sect.D V. 63 1243 2007.
ISSN: ISSN 0907-4449
PubMed: 18084072
DOI: 10.1107/S0907444907052985
Page generated: Wed Aug 20 05:23:23 2025
ISSN: ISSN 0907-4449
PubMed: 18084072
DOI: 10.1107/S0907444907052985
Last articles
Zn in 3HK8Zn in 3HK5
Zn in 3HJW
Zn in 3HCS
Zn in 3HJT
Zn in 3HI2
Zn in 3HGZ
Zn in 3HFF
Zn in 3HFY
Zn in 3HCU