Zinc »
PDB 2kzz-2lgl »
2l75 »
Zinc in PDB 2l75: Solution Structure of CHD4-PHD2 in Complex with H3K9ME3
Zinc Binding Sites:
The binding sites of Zinc atom in the Solution Structure of CHD4-PHD2 in Complex with H3K9ME3
(pdb code 2l75). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Solution Structure of CHD4-PHD2 in Complex with H3K9ME3, PDB code: 2l75:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Solution Structure of CHD4-PHD2 in Complex with H3K9ME3, PDB code: 2l75:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 2l75
Go back to
Zinc binding site 1 out
of 2 in the Solution Structure of CHD4-PHD2 in Complex with H3K9ME3
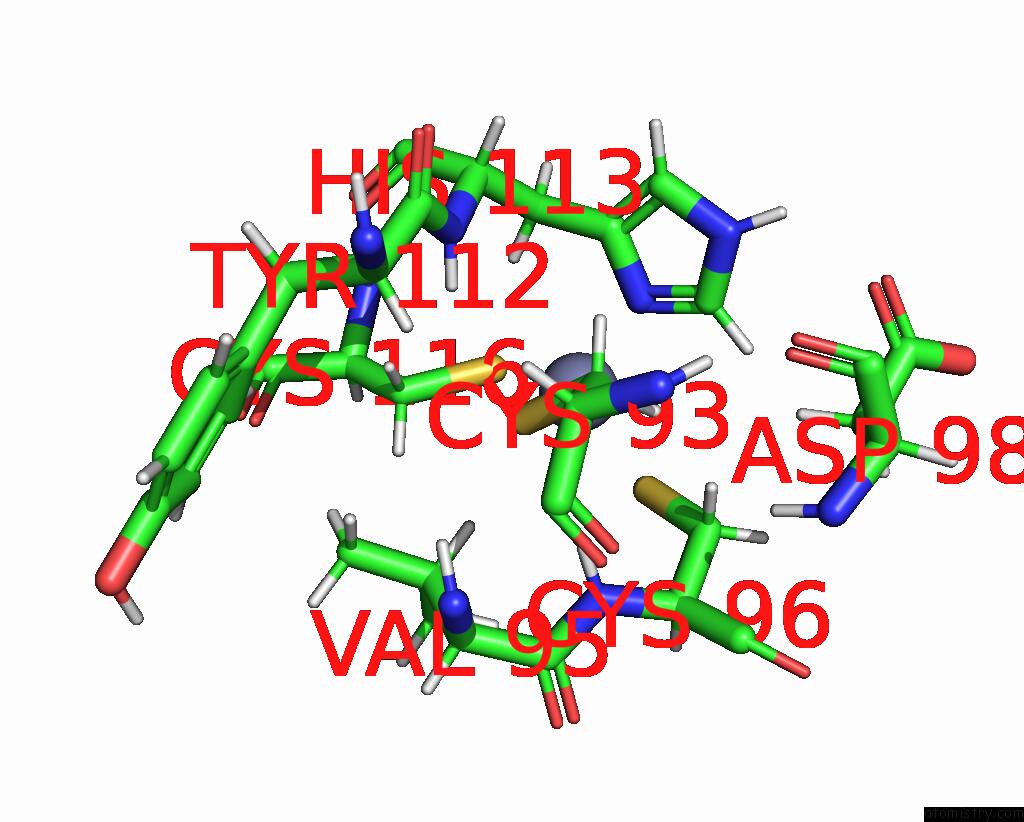
Mono view
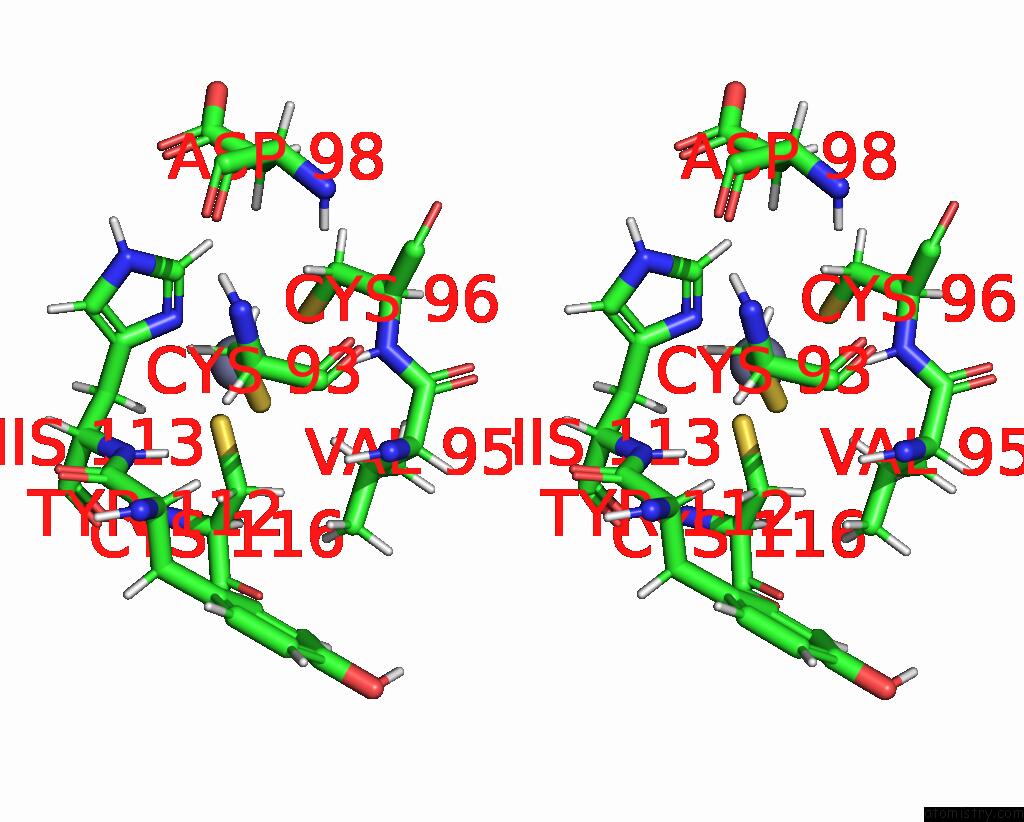
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution Structure of CHD4-PHD2 in Complex with H3K9ME3 within 5.0Å range:
|
Zinc binding site 2 out of 2 in 2l75
Go back to
Zinc binding site 2 out
of 2 in the Solution Structure of CHD4-PHD2 in Complex with H3K9ME3
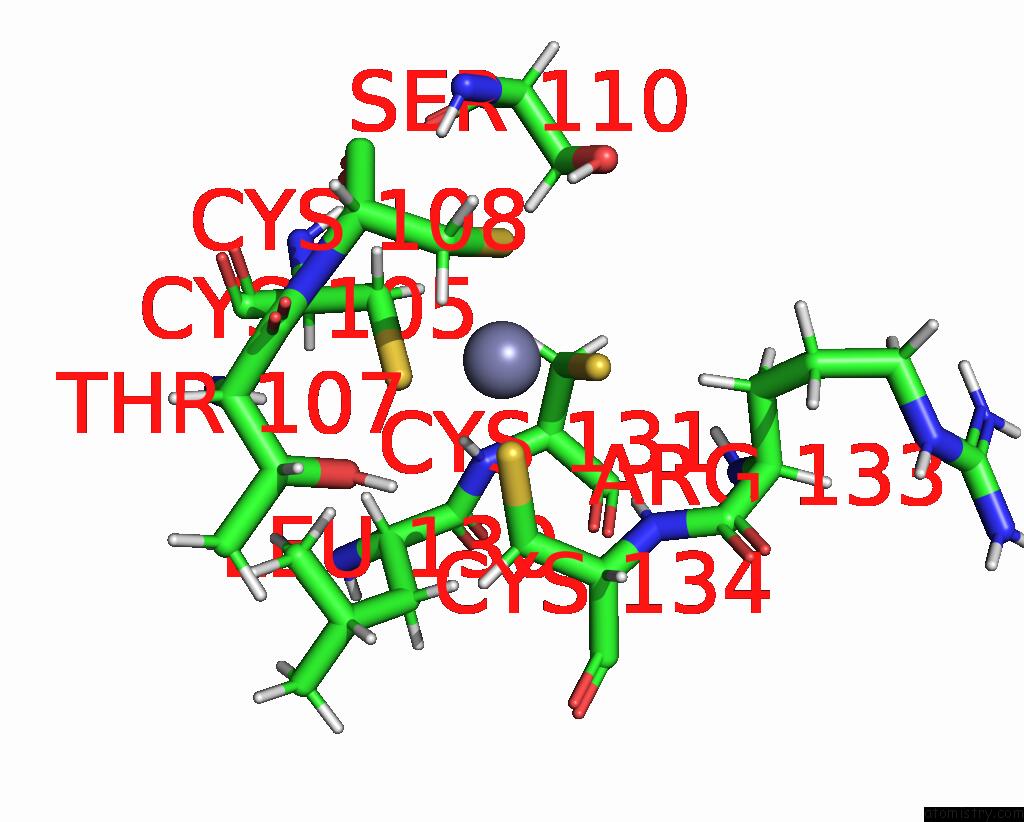
Mono view
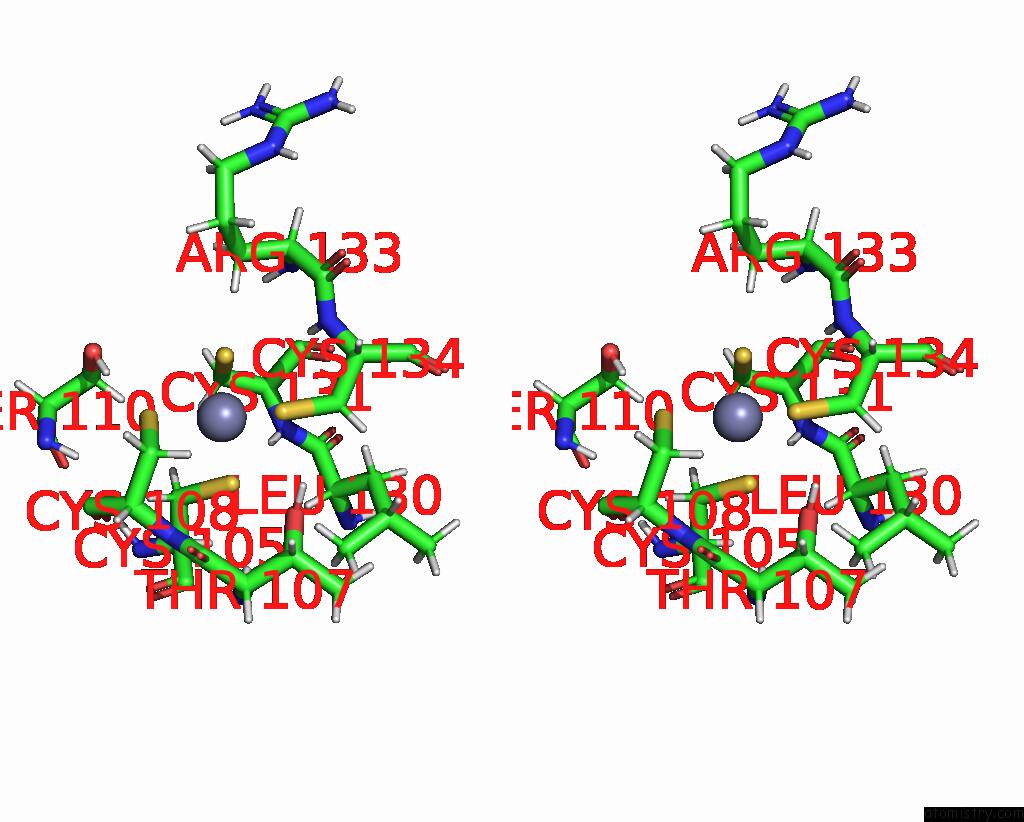
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Solution Structure of CHD4-PHD2 in Complex with H3K9ME3 within 5.0Å range:
|
Reference:
R.E.Mansfield,
C.A.Musselman,
A.H.Kwan,
S.S.Oliver,
A.L.Garske,
F.Davrazou,
J.M.Denu,
T.G.Kutateladze,
J.P.Mackay.
Plant Homeodomain (Phd) Fingers of CHD4 Are Histone H3-Binding Modules with Preference For Unmodified H3K4 and Methylated H3K9 J.Biol.Chem. V. 286 11779 2011.
ISSN: ISSN 0021-9258
PubMed: 21278251
DOI: 10.1074/JBC.M110.208207
Page generated: Thu Oct 17 01:46:32 2024
ISSN: ISSN 0021-9258
PubMed: 21278251
DOI: 10.1074/JBC.M110.208207
Last articles
As in 2G6OAs in 2C7V
As in 2CJW
As in 2E11
As in 2FAU
As in 2DYB
As in 2D28
As in 2BVT
As in 2BVY
As in 2BOX