Zinc »
PDB 2jun-2kem »
2k2c »
Zinc in PDB 2k2c: Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A
Zinc Binding Sites:
The binding sites of Zinc atom in the Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A
(pdb code 2k2c). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 6 binding sites of Zinc where determined in the Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A, PDB code: 2k2c:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6;
In total 6 binding sites of Zinc where determined in the Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A, PDB code: 2k2c:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6;
Zinc binding site 1 out of 6 in 2k2c
Go back to
Zinc binding site 1 out
of 6 in the Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A within 5.0Å range:
|
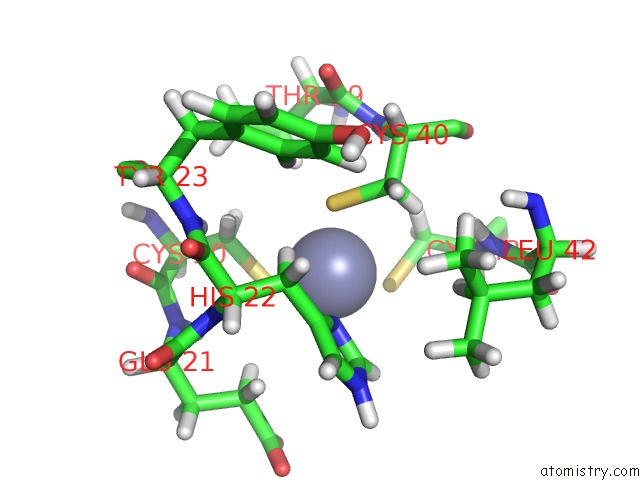
Zinc binding site 2 out of 6 in 2k2c
Go back to
Zinc binding site 2 out
of 6 in the Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A
Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A within 5.0Å range:
|
Zinc binding site 3 out of 6 in 2k2c
Go back to
Zinc binding site 3 out
of 6 in the Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A within 5.0Å range:
|
Zinc binding site 4 out of 6 in 2k2c
Go back to
Zinc binding site 4 out
of 6 in the Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A

Mono view

Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A within 5.0Å range:
|
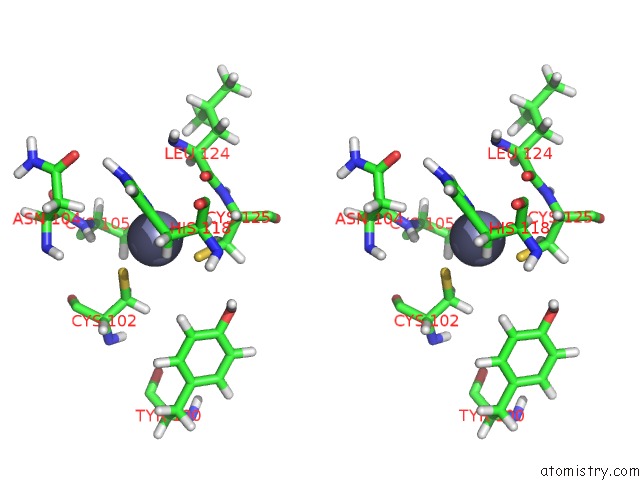
Zinc binding site 5 out of 6 in 2k2c
Go back to
Zinc binding site 5 out
of 6 in the Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A within 5.0Å range:
|
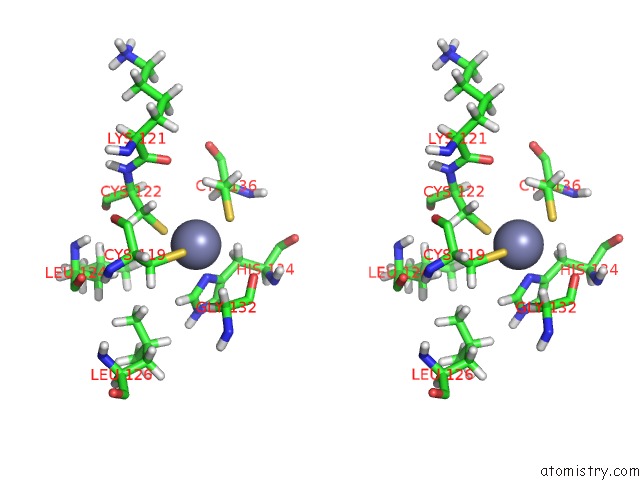
Zinc binding site 6 out of 6 in 2k2c
Go back to
Zinc binding site 6 out
of 6 in the Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A

Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Solution uc(Nmr) Structure of N-Terminal Domain of Human PIRH2. Northeast Structural Genomics Consortium (Nesg) Target HT2A within 5.0Å range:
|
Reference:
Y.Sheng,
R.C.Laister,
A.Lemak,
B.Wu,
E.Tai,
S.Duan,
J.Lukin,
M.Sunnerhagen,
S.Srisailam,
M.Karra,
S.Benchimol,
C.H.Arrowsmith.
Molecular Basis of PIRH2-Mediated P53 Ubiquitylation. Nat.Struct.Mol.Biol. V. 15 1334 2008.
ISSN: ISSN 1545-9993
PubMed: 19043414
DOI: 10.1038/NSMB.1521
Page generated: Thu Oct 17 01:30:14 2024
ISSN: ISSN 1545-9993
PubMed: 19043414
DOI: 10.1038/NSMB.1521
Last articles
Ag in 7ECNAg in 7CGC
Ag in 7DC1
Ag in 7CB6
Ag in 7BSH
Ag in 7BSE
Ag in 6M2P
Ag in 7C17
Ag in 7BSF
Ag in 7BSG