Zinc »
PDB 2jun-2kem »
2k0a »
Zinc in PDB 2k0a: 1H, 15N and 13C Chemical Shift Assignments For RDS3 Protein
Zinc Binding Sites:
The binding sites of Zinc atom in the 1H, 15N and 13C Chemical Shift Assignments For RDS3 Protein
(pdb code 2k0a). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 3 binding sites of Zinc where determined in the 1H, 15N and 13C Chemical Shift Assignments For RDS3 Protein, PDB code: 2k0a:
Jump to Zinc binding site number: 1; 2; 3;
In total 3 binding sites of Zinc where determined in the 1H, 15N and 13C Chemical Shift Assignments For RDS3 Protein, PDB code: 2k0a:
Jump to Zinc binding site number: 1; 2; 3;
Zinc binding site 1 out of 3 in 2k0a
Go back to
Zinc binding site 1 out
of 3 in the 1H, 15N and 13C Chemical Shift Assignments For RDS3 Protein
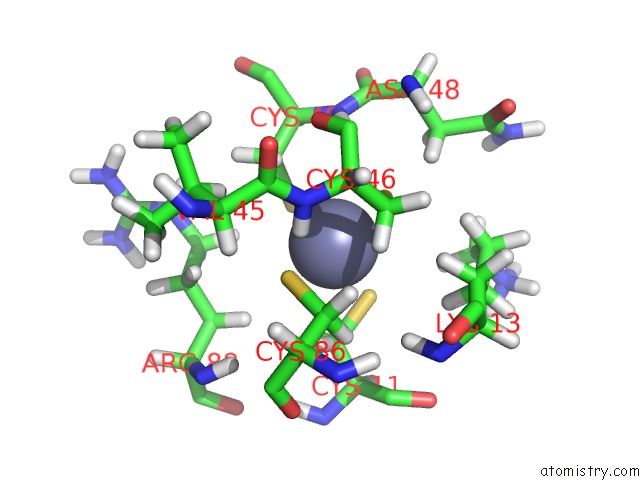
Mono view
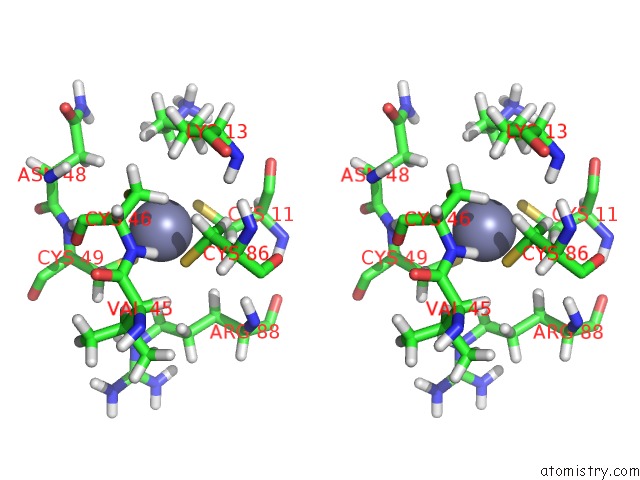
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of 1H, 15N and 13C Chemical Shift Assignments For RDS3 Protein within 5.0Å range:
|
Zinc binding site 2 out of 3 in 2k0a
Go back to
Zinc binding site 2 out
of 3 in the 1H, 15N and 13C Chemical Shift Assignments For RDS3 Protein
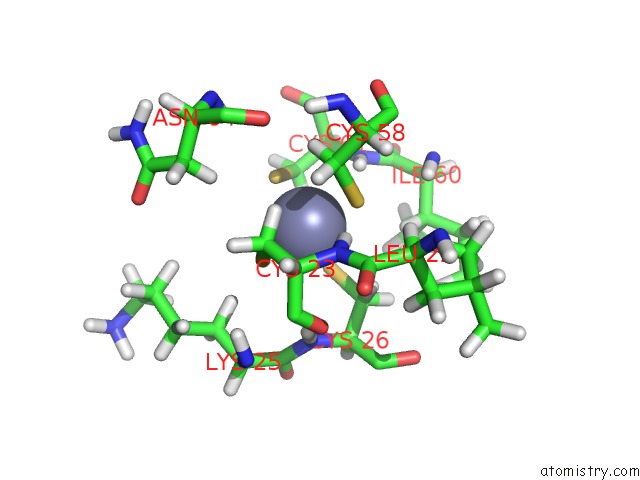
Mono view
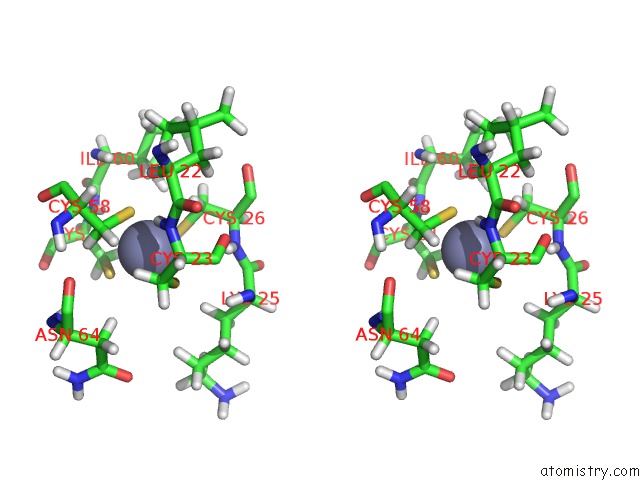
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of 1H, 15N and 13C Chemical Shift Assignments For RDS3 Protein within 5.0Å range:
|
Zinc binding site 3 out of 3 in 2k0a
Go back to
Zinc binding site 3 out
of 3 in the 1H, 15N and 13C Chemical Shift Assignments For RDS3 Protein
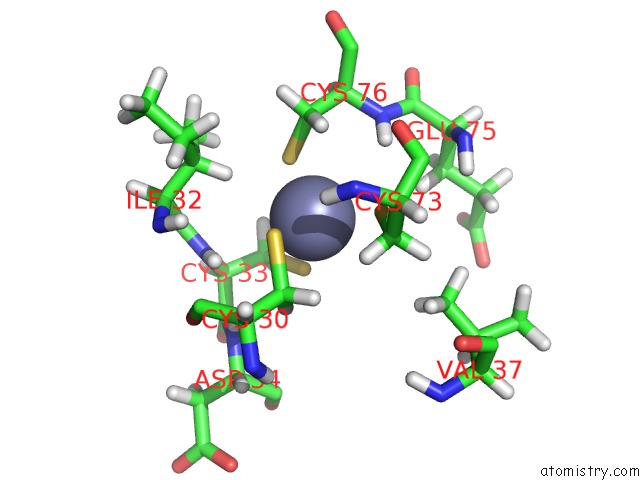
Mono view
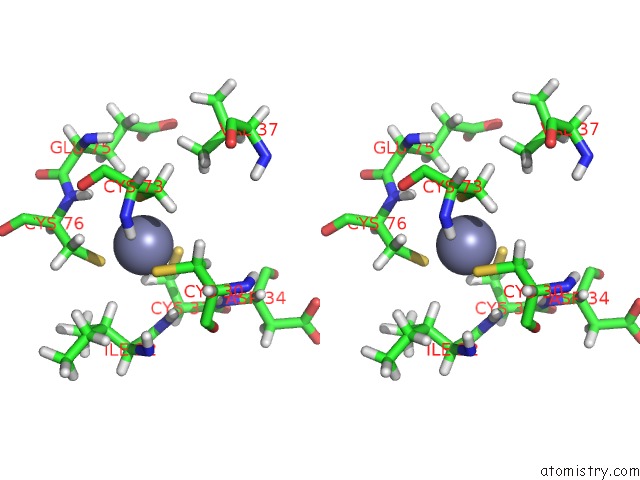
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of 1H, 15N and 13C Chemical Shift Assignments For RDS3 Protein within 5.0Å range:
|
Reference:
A.M.Van Roon,
N.M.Loening,
E.Obayashi,
J.C.Yang,
A.J.Newman,
H.Hernandez,
K.Nagai,
D.Neuhaus.
Solution Structure of the U2 Snrnp Protein RDS3P Reveals A Knotted Zinc-Finger Motif. Proc.Natl.Acad.Sci.Usa V. 105 9621 2008.
ISSN: ISSN 0027-8424
PubMed: 18621724
DOI: 10.1073/PNAS.0802494105
Page generated: Thu Oct 17 01:28:47 2024
ISSN: ISSN 0027-8424
PubMed: 18621724
DOI: 10.1073/PNAS.0802494105
Last articles
Ag in 7SDTAg in 7ECL
Ag in 7SDS
Ag in 7SDQ
Ag in 7SDO
Ag in 7SDN
Ag in 7SDM
Ag in 7SDL
Ag in 7SDK
Ag in 7SDG