Zinc »
PDB 2jb0-2jtn »
2jmi »
Zinc in PDB 2jmi: uc(Nmr) Solution Structure of Phd Finger Fragment of Yeast YNG1 Protein in Free State
Zinc Binding Sites:
The binding sites of Zinc atom in the uc(Nmr) Solution Structure of Phd Finger Fragment of Yeast YNG1 Protein in Free State
(pdb code 2jmi). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the uc(Nmr) Solution Structure of Phd Finger Fragment of Yeast YNG1 Protein in Free State, PDB code: 2jmi:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the uc(Nmr) Solution Structure of Phd Finger Fragment of Yeast YNG1 Protein in Free State, PDB code: 2jmi:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 2jmi
Go back to
Zinc binding site 1 out
of 2 in the uc(Nmr) Solution Structure of Phd Finger Fragment of Yeast YNG1 Protein in Free State
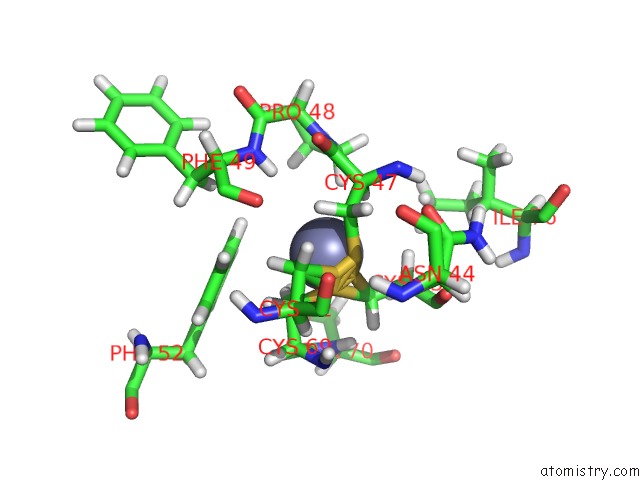
Mono view
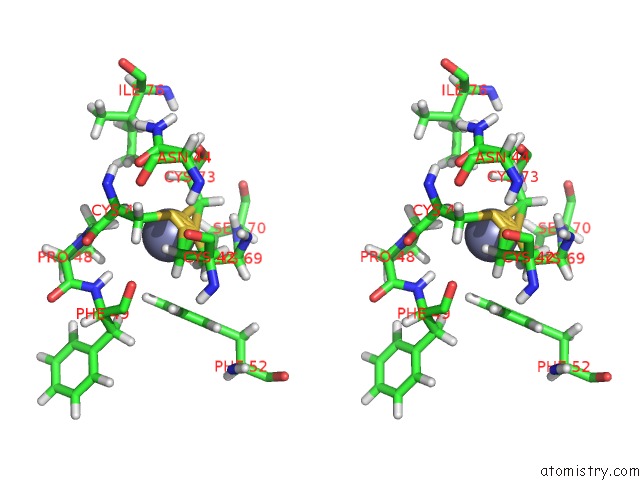
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of uc(Nmr) Solution Structure of Phd Finger Fragment of Yeast YNG1 Protein in Free State within 5.0Å range:
|
Zinc binding site 2 out of 2 in 2jmi
Go back to
Zinc binding site 2 out
of 2 in the uc(Nmr) Solution Structure of Phd Finger Fragment of Yeast YNG1 Protein in Free State
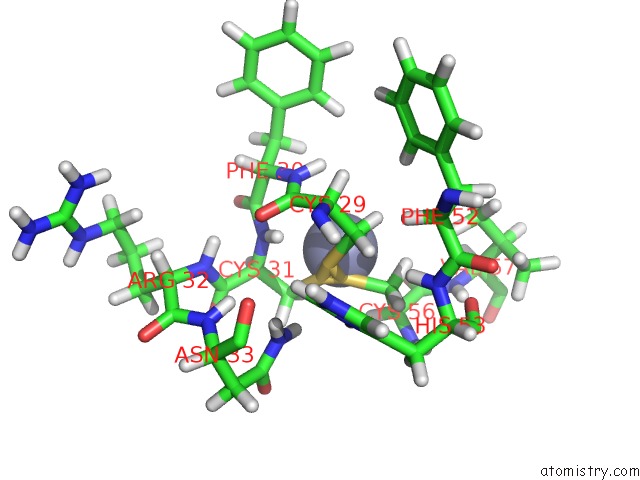
Mono view
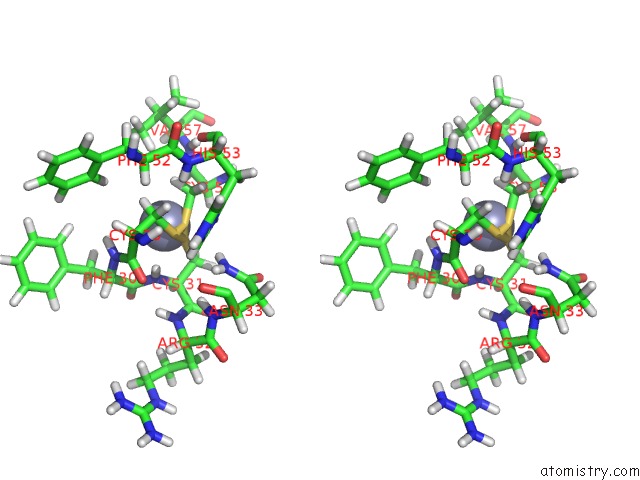
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of uc(Nmr) Solution Structure of Phd Finger Fragment of Yeast YNG1 Protein in Free State within 5.0Å range:
|
Reference:
S.D.Taverna,
S.Ilin,
R.S.Rogers,
J.C.Tanny,
H.Lavender,
H.Li,
L.Baker,
J.Boyle,
L.P.Blair,
B.T.Chait,
D.J.Patel,
J.D.Aitchison,
A.J.Tackett,
C.D.Allis.
YNG1 Phd Finger Binding to H3 Trimethylated at K4 Promotes NUA3 Hat Activity at K14 of H3 and Transcription at A Subset of Targeted Orfs Mol.Cell V. 24 785 2006.
ISSN: ISSN 1097-2765
PubMed: 17157260
DOI: 10.1016/J.MOLCEL.2006.10.026
Page generated: Thu Oct 17 01:17:29 2024
ISSN: ISSN 1097-2765
PubMed: 17157260
DOI: 10.1016/J.MOLCEL.2006.10.026
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF