Zinc »
PDB 2jb0-2jtn »
2jm1 »
Zinc in PDB 2jm1: Structures and Chemical Shift Assignments For the Add Domain of the Atrx Protein
Zinc Binding Sites:
The binding sites of Zinc atom in the Structures and Chemical Shift Assignments For the Add Domain of the Atrx Protein
(pdb code 2jm1). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 3 binding sites of Zinc where determined in the Structures and Chemical Shift Assignments For the Add Domain of the Atrx Protein, PDB code: 2jm1:
Jump to Zinc binding site number: 1; 2; 3;
In total 3 binding sites of Zinc where determined in the Structures and Chemical Shift Assignments For the Add Domain of the Atrx Protein, PDB code: 2jm1:
Jump to Zinc binding site number: 1; 2; 3;
Zinc binding site 1 out of 3 in 2jm1
Go back to
Zinc binding site 1 out
of 3 in the Structures and Chemical Shift Assignments For the Add Domain of the Atrx Protein
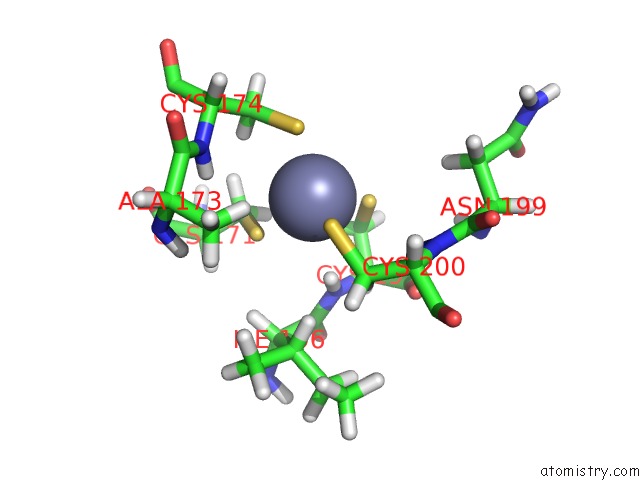
Mono view
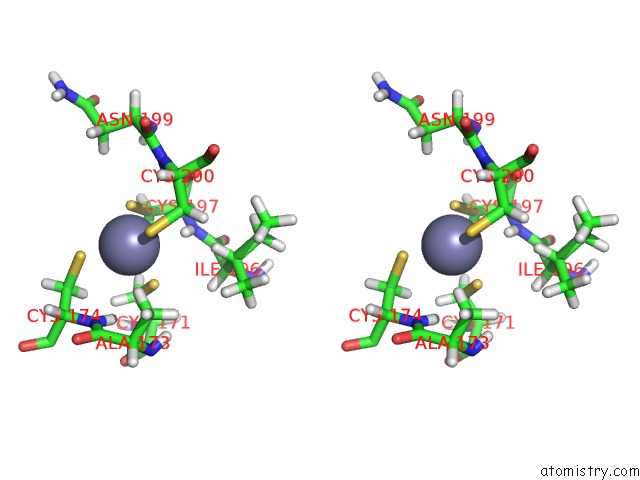
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Structures and Chemical Shift Assignments For the Add Domain of the Atrx Protein within 5.0Å range:
|
Zinc binding site 2 out of 3 in 2jm1
Go back to
Zinc binding site 2 out
of 3 in the Structures and Chemical Shift Assignments For the Add Domain of the Atrx Protein
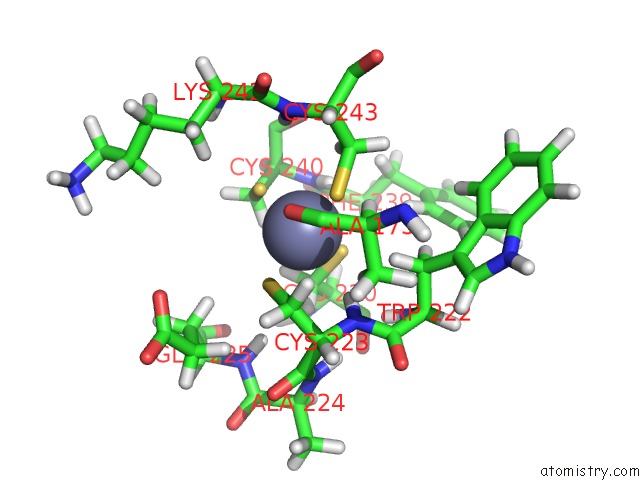
Mono view
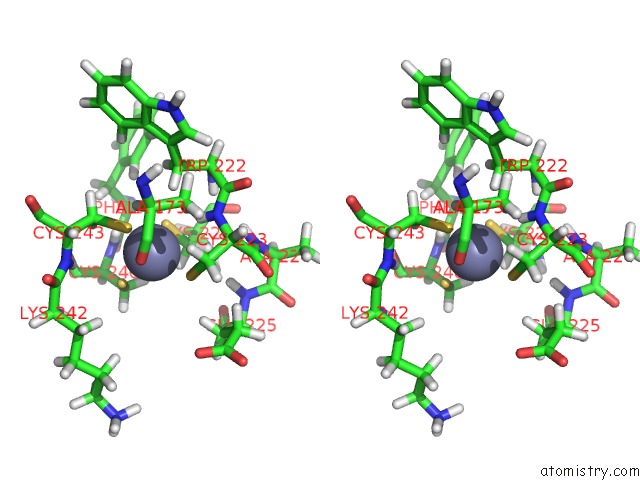
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Structures and Chemical Shift Assignments For the Add Domain of the Atrx Protein within 5.0Å range:
|
Zinc binding site 3 out of 3 in 2jm1
Go back to
Zinc binding site 3 out
of 3 in the Structures and Chemical Shift Assignments For the Add Domain of the Atrx Protein
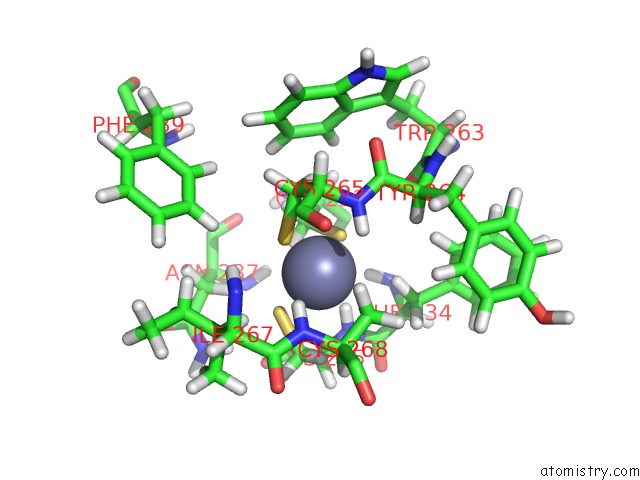
Mono view
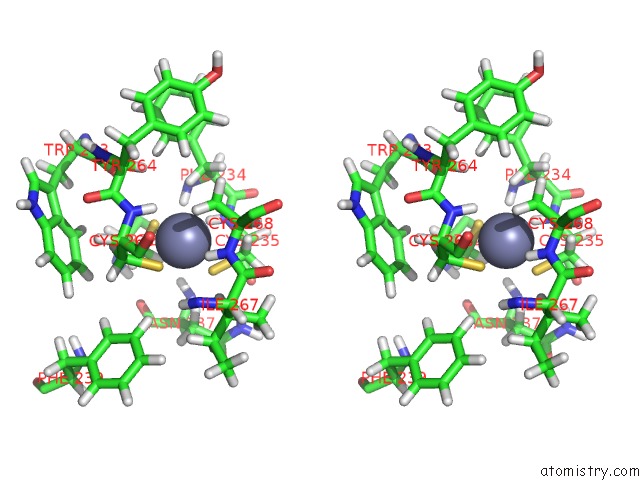
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Structures and Chemical Shift Assignments For the Add Domain of the Atrx Protein within 5.0Å range:
|
Reference:
A.Argentaro,
J.C.Yang,
L.Chapman,
M.S.Kowalczyk,
R.J.Gibbons,
D.R.Higgs,
D.Neuhaus,
D.Rhodes.
Structural Consequences of Disease-Causing Mutations in the Atrx-DNMT3-DNMT3L (Add) Domain of the Chromatin-Associated Protein Atrx. Proc.Natl.Acad.Sci.Usa V. 104 11939 2007.
ISSN: ISSN 0027-8424
PubMed: 17609377
DOI: 10.1073/PNAS.0704057104
Page generated: Wed Aug 20 03:49:45 2025
ISSN: ISSN 0027-8424
PubMed: 17609377
DOI: 10.1073/PNAS.0704057104
Last articles
Zn in 3DGDZn in 3DGN
Zn in 3DDT
Zn in 3DGM
Zn in 3DFM
Zn in 3DGL
Zn in 3DFX
Zn in 3DFV
Zn in 3DFK
Zn in 3DFF