Zinc »
PDB 2fos-2g2p »
2fuu »
Zinc in PDB 2fuu: uc(Nmr) Solution Structure of the Phd Domain From the Human Bptf in Complex with H3(1-15)K4ME3 Peptide
Zinc Binding Sites:
The binding sites of Zinc atom in the uc(Nmr) Solution Structure of the Phd Domain From the Human Bptf in Complex with H3(1-15)K4ME3 Peptide
(pdb code 2fuu). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the uc(Nmr) Solution Structure of the Phd Domain From the Human Bptf in Complex with H3(1-15)K4ME3 Peptide, PDB code: 2fuu:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the uc(Nmr) Solution Structure of the Phd Domain From the Human Bptf in Complex with H3(1-15)K4ME3 Peptide, PDB code: 2fuu:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 2fuu
Go back to
Zinc binding site 1 out
of 2 in the uc(Nmr) Solution Structure of the Phd Domain From the Human Bptf in Complex with H3(1-15)K4ME3 Peptide
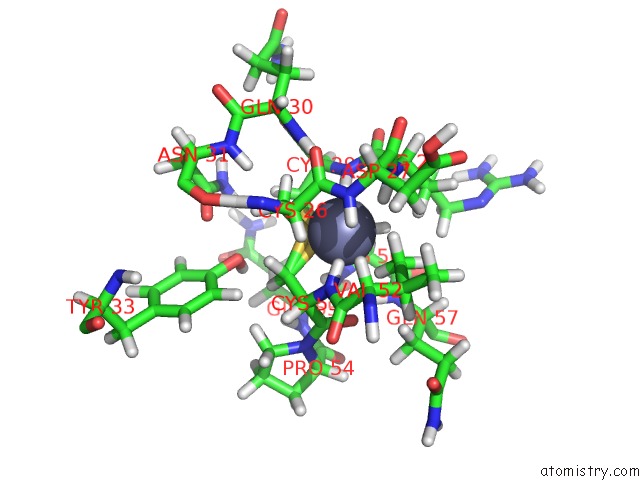
Mono view
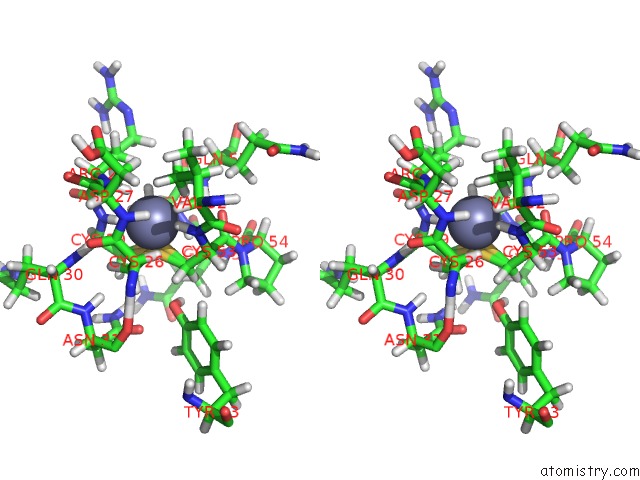
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of uc(Nmr) Solution Structure of the Phd Domain From the Human Bptf in Complex with H3(1-15)K4ME3 Peptide within 5.0Å range:
|
Zinc binding site 2 out of 2 in 2fuu
Go back to
Zinc binding site 2 out
of 2 in the uc(Nmr) Solution Structure of the Phd Domain From the Human Bptf in Complex with H3(1-15)K4ME3 Peptide
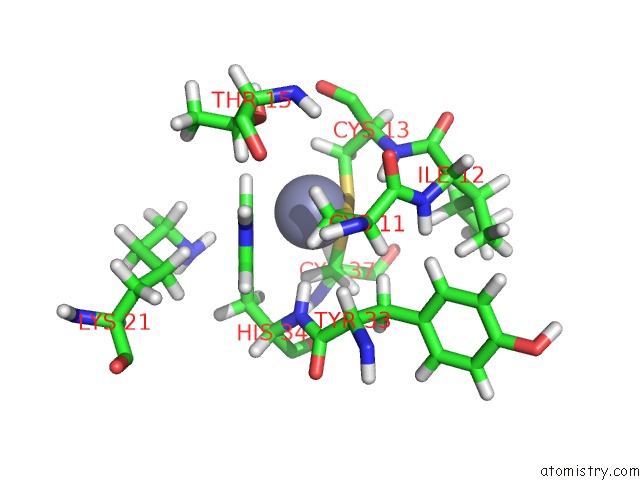
Mono view
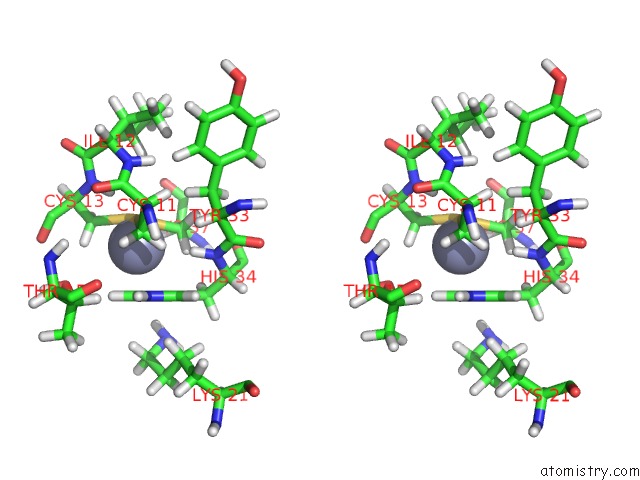
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of uc(Nmr) Solution Structure of the Phd Domain From the Human Bptf in Complex with H3(1-15)K4ME3 Peptide within 5.0Å range:
|
Reference:
H.Li,
S.Ilin,
W.Wang,
E.M.Duncan,
J.Wysocka,
C.D.Allis,
D.J.Patel.
Molecular Basis For Site-Specific Read-Out of Histone H3K4ME3 By the Bptf Phd Finger of Nurf. Nature V. 442 91 2006.
ISSN: ISSN 0028-0836
PubMed: 16728978
DOI: 10.1038/NATURE04802
Page generated: Wed Oct 16 23:55:32 2024
ISSN: ISSN 0028-0836
PubMed: 16728978
DOI: 10.1038/NATURE04802
Last articles
Zn in 9MJ5Zn in 9HNW
Zn in 9G0L
Zn in 9FNE
Zn in 9DZN
Zn in 9E0I
Zn in 9D32
Zn in 9DAK
Zn in 8ZXC
Zn in 8ZUF