Zinc »
PDB 2e3x-2eer »
2ea0 »
Zinc in PDB 2ea0: Crystal Structure of the Dna Repair Enzyme Endonuclease-VIII (Nei) From E. Coli in Complex with Ap-Site Containing Dna Substrate
Enzymatic activity of Crystal Structure of the Dna Repair Enzyme Endonuclease-VIII (Nei) From E. Coli in Complex with Ap-Site Containing Dna Substrate
All present enzymatic activity of Crystal Structure of the Dna Repair Enzyme Endonuclease-VIII (Nei) From E. Coli in Complex with Ap-Site Containing Dna Substrate:
4.2.99.18;
4.2.99.18;
Protein crystallography data
The structure of Crystal Structure of the Dna Repair Enzyme Endonuclease-VIII (Nei) From E. Coli in Complex with Ap-Site Containing Dna Substrate, PDB code: 2ea0
was solved by
G.Golan,
D.O.Zharov,
A.P.Grollman,
G.Shoham,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 30.00 / 1.40 |
| Space group | P 43 2 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 76.184, 76.184, 164.540, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 15.6 / 20.4 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of the Dna Repair Enzyme Endonuclease-VIII (Nei) From E. Coli in Complex with Ap-Site Containing Dna Substrate
(pdb code 2ea0). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Crystal Structure of the Dna Repair Enzyme Endonuclease-VIII (Nei) From E. Coli in Complex with Ap-Site Containing Dna Substrate, PDB code: 2ea0:
In total only one binding site of Zinc was determined in the Crystal Structure of the Dna Repair Enzyme Endonuclease-VIII (Nei) From E. Coli in Complex with Ap-Site Containing Dna Substrate, PDB code: 2ea0:
Zinc binding site 1 out of 1 in 2ea0
Go back to
Zinc binding site 1 out
of 1 in the Crystal Structure of the Dna Repair Enzyme Endonuclease-VIII (Nei) From E. Coli in Complex with Ap-Site Containing Dna Substrate
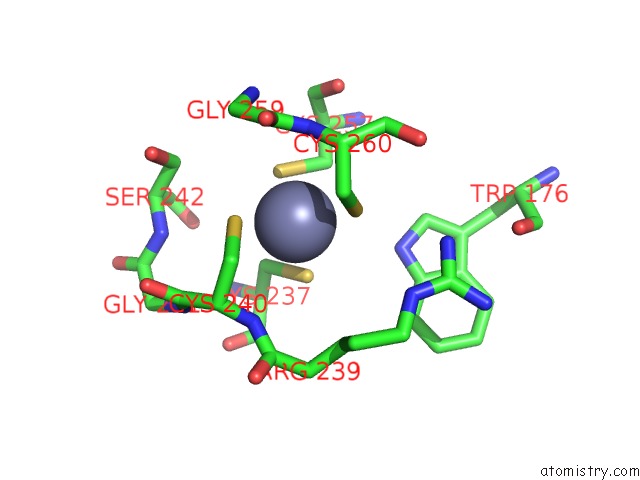
Mono view
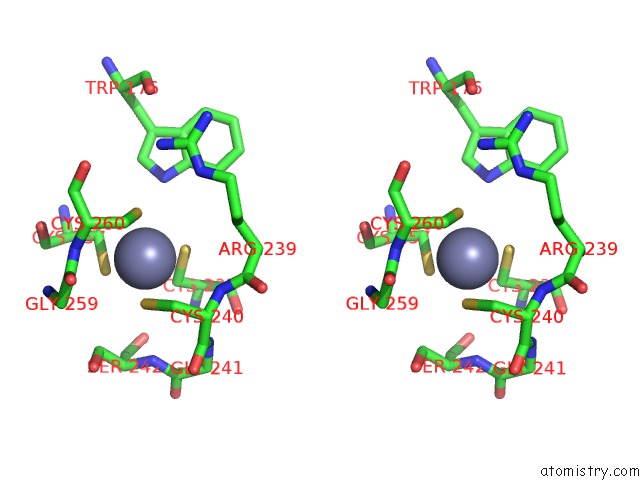
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of the Dna Repair Enzyme Endonuclease-VIII (Nei) From E. Coli in Complex with Ap-Site Containing Dna Substrate within 5.0Å range:
|
Reference:
G.Golan,
D.O.Zharkov,
A.P.Grollman,
G.Shoham.
Active Site Plasticity of Endonuclease VIII: Conformational Changes Compensating For Different Substrate and Mutations of Critical Residues To Be Published.
Page generated: Wed Aug 20 02:12:10 2025
Last articles
Zn in 2XJLZn in 2XJK
Zn in 2XDD
Zn in 2XEV
Zn in 2XHM
Zn in 2XGW
Zn in 2XHL
Zn in 2XF4
Zn in 2XEU
Zn in 2XEJ