Zinc »
PDB 1xv2-1y8j »
1y0j »
Zinc in PDB 1y0j: Zinc Fingers As Protein Recognition Motifs: Structural Basis For the Gata-1/Friend of Gata Interaction
Zinc Binding Sites:
The binding sites of Zinc atom in the Zinc Fingers As Protein Recognition Motifs: Structural Basis For the Gata-1/Friend of Gata Interaction
(pdb code 1y0j). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Zinc Fingers As Protein Recognition Motifs: Structural Basis For the Gata-1/Friend of Gata Interaction, PDB code: 1y0j:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Zinc Fingers As Protein Recognition Motifs: Structural Basis For the Gata-1/Friend of Gata Interaction, PDB code: 1y0j:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 1y0j
Go back to
Zinc binding site 1 out
of 2 in the Zinc Fingers As Protein Recognition Motifs: Structural Basis For the Gata-1/Friend of Gata Interaction
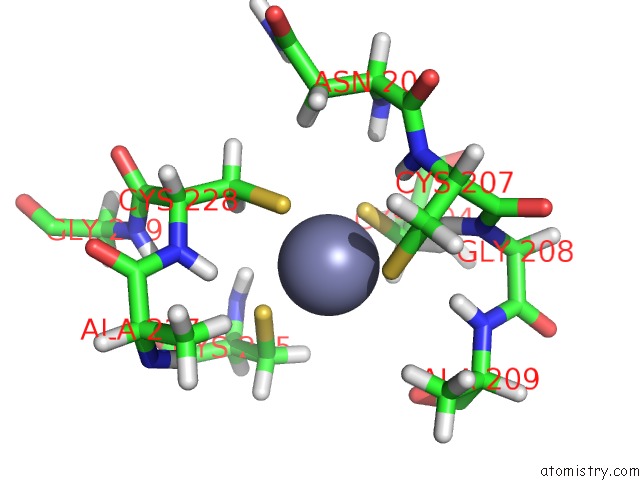
Mono view
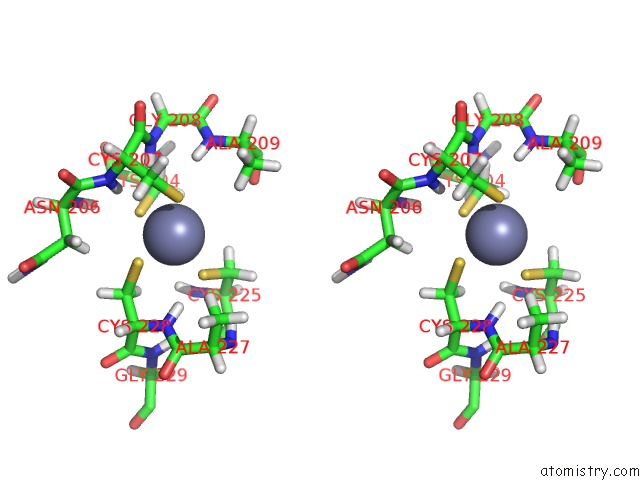
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Zinc Fingers As Protein Recognition Motifs: Structural Basis For the Gata-1/Friend of Gata Interaction within 5.0Å range:
|
Zinc binding site 2 out of 2 in 1y0j
Go back to
Zinc binding site 2 out
of 2 in the Zinc Fingers As Protein Recognition Motifs: Structural Basis For the Gata-1/Friend of Gata Interaction
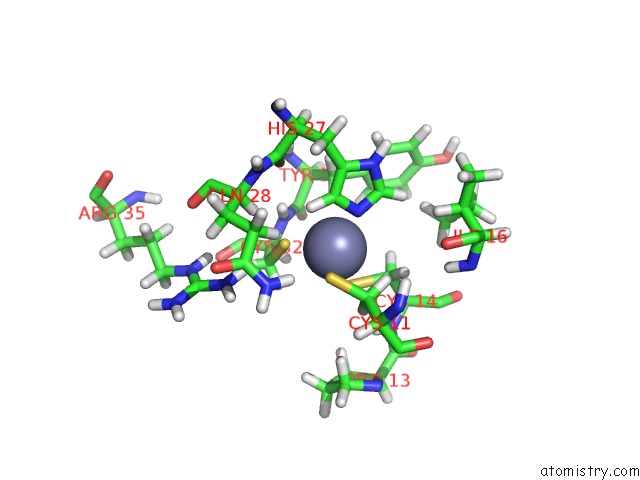
Mono view
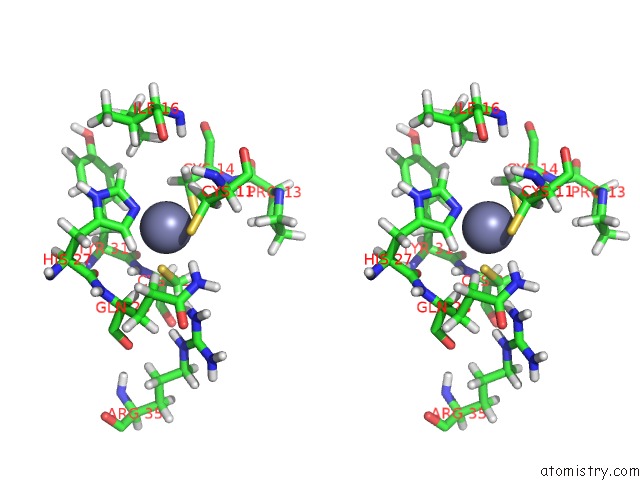
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Zinc Fingers As Protein Recognition Motifs: Structural Basis For the Gata-1/Friend of Gata Interaction within 5.0Å range:
|
Reference:
C.K.Liew,
R.J.Y.Simpson,
A.H.Y.Kwan,
L.A.Crofts,
F.E.Loughlin,
J.M.Matthews,
M.Crossley,
J.P.Mackay.
Zinc Fingers As Protein Recognition Motifs: Structural Basis For the Gata-1/Friend of Gata Interaction Proc.Natl.Acad.Sci.Usa V. 102 583 2005.
ISSN: ISSN 0027-8424
PubMed: 15644435
DOI: 10.1073/PNAS.0407511102
Page generated: Wed Aug 20 00:28:48 2025
ISSN: ISSN 0027-8424
PubMed: 15644435
DOI: 10.1073/PNAS.0407511102
Last articles
Zn in 2GZGZn in 2GZF
Zn in 2GZE
Zn in 2GVI
Zn in 2GYK
Zn in 2GWN
Zn in 2GWG
Zn in 2GVF
Zn in 2GVM
Zn in 2GU2