Zinc »
PDB 1wwe-1x8g »
1x6m »
Zinc in PDB 1x6m: Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa)
Protein crystallography data
The structure of Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa), PDB code: 1x6m
was solved by
A.M.Neculai,
D.Neculai,
J.A.Vorholt,
S.Becker,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 74.29 / 2.35 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 53.360, 118.860, 95.990, 90.00, 97.48, 90.00 |
| R / Rfree (%) | 18.9 / 25 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa)
(pdb code 1x6m). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 8 binding sites of Zinc where determined in the Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa), PDB code: 1x6m:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Zinc where determined in the Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa), PDB code: 1x6m:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Zinc binding site 1 out of 8 in 1x6m
Go back to
Zinc binding site 1 out
of 8 in the Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa)
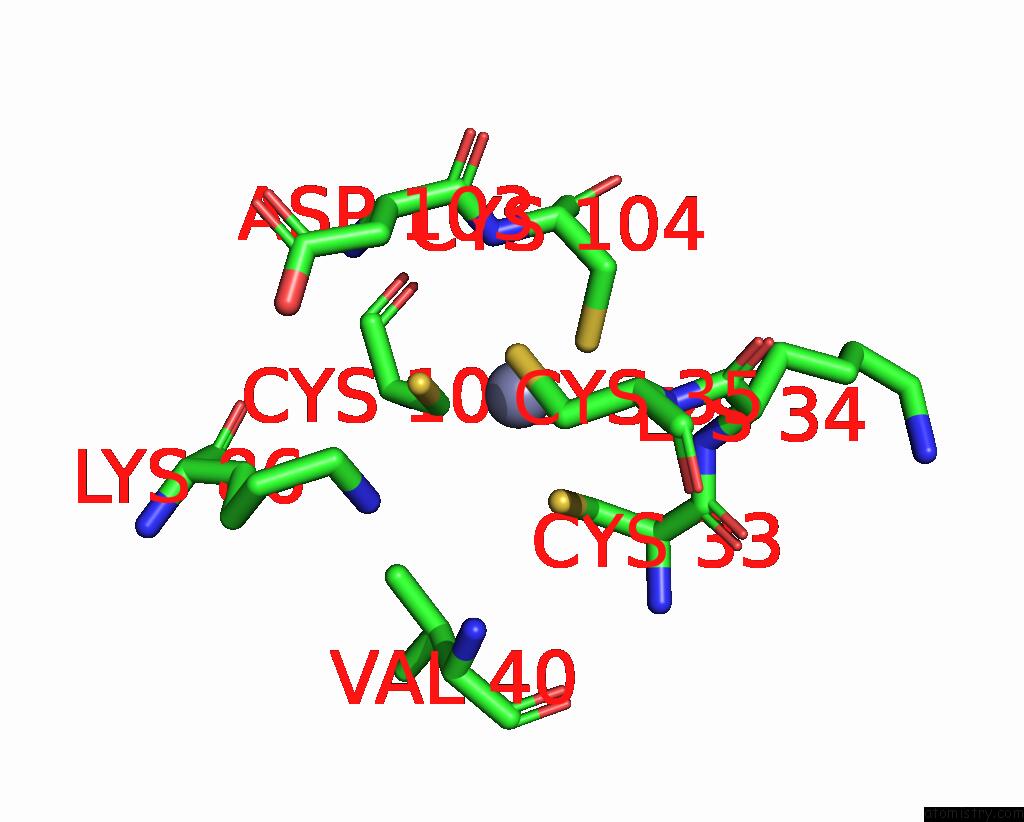
Mono view
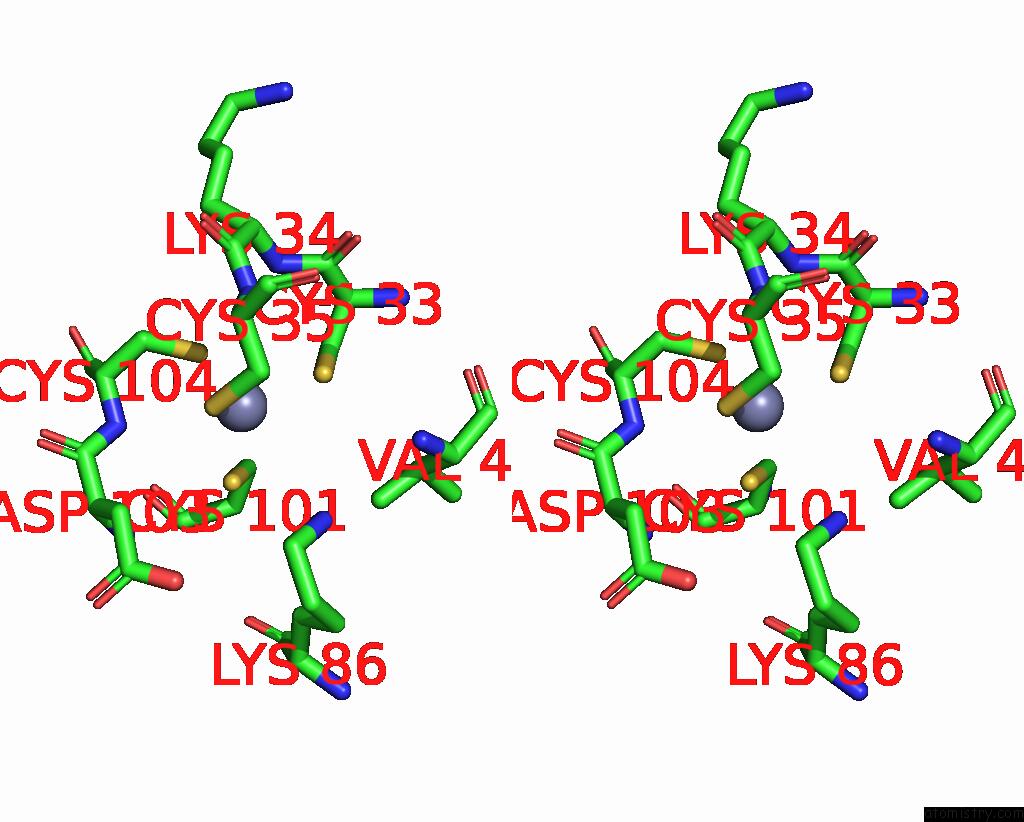
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa) within 5.0Å range:
|
Zinc binding site 2 out of 8 in 1x6m
Go back to
Zinc binding site 2 out
of 8 in the Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa)
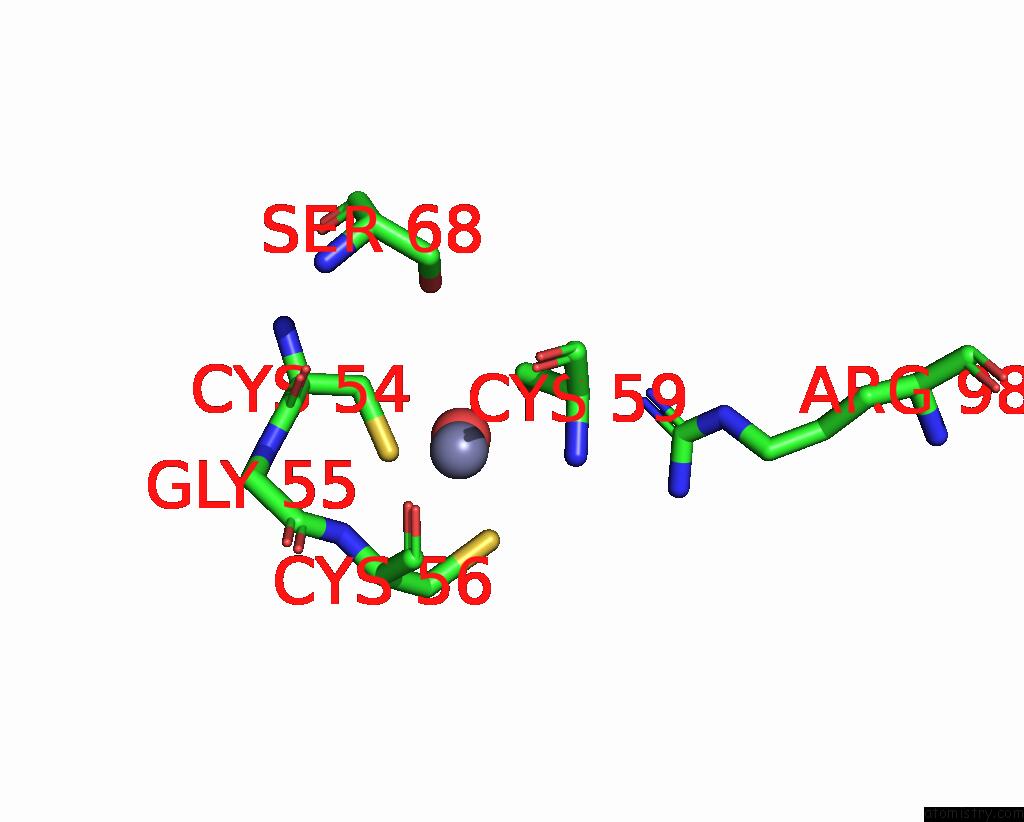
Mono view
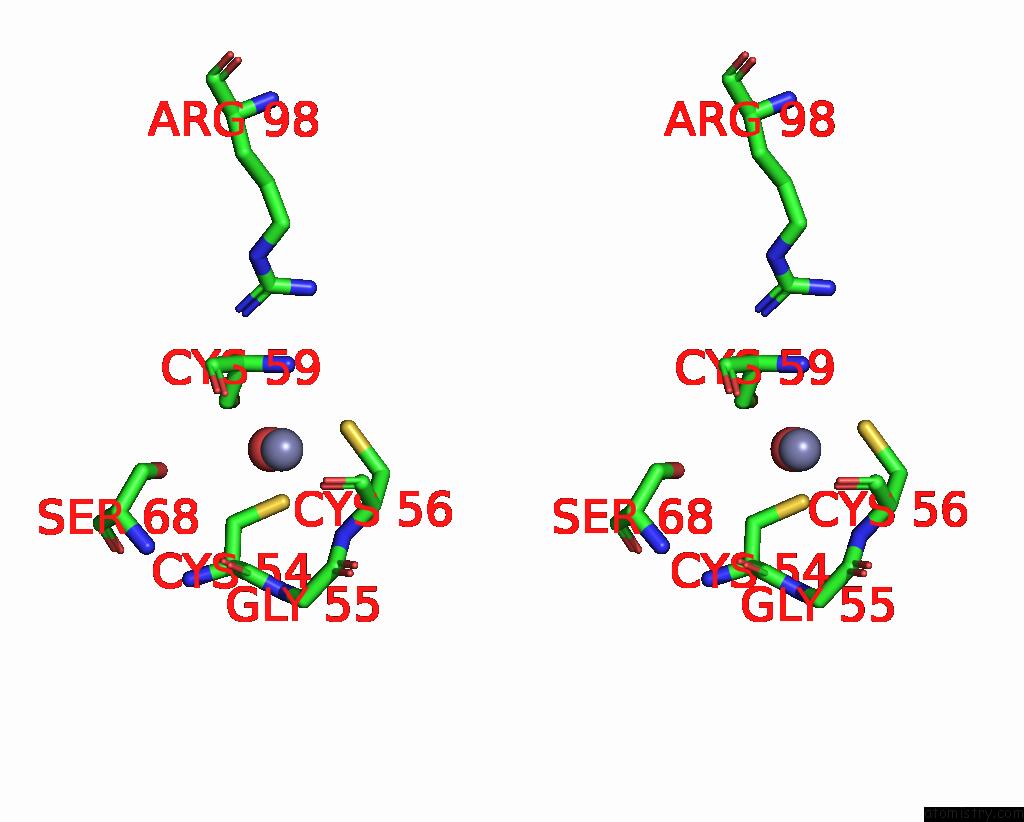
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa) within 5.0Å range:
|
Zinc binding site 3 out of 8 in 1x6m
Go back to
Zinc binding site 3 out
of 8 in the Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa)
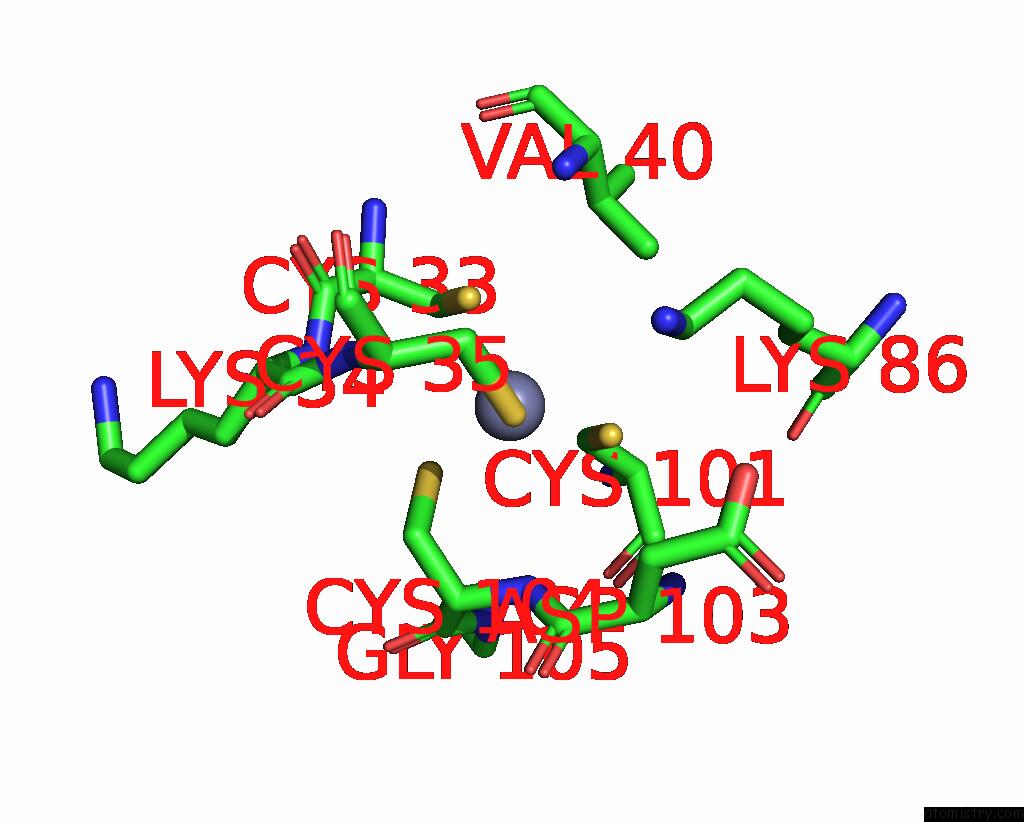
Mono view
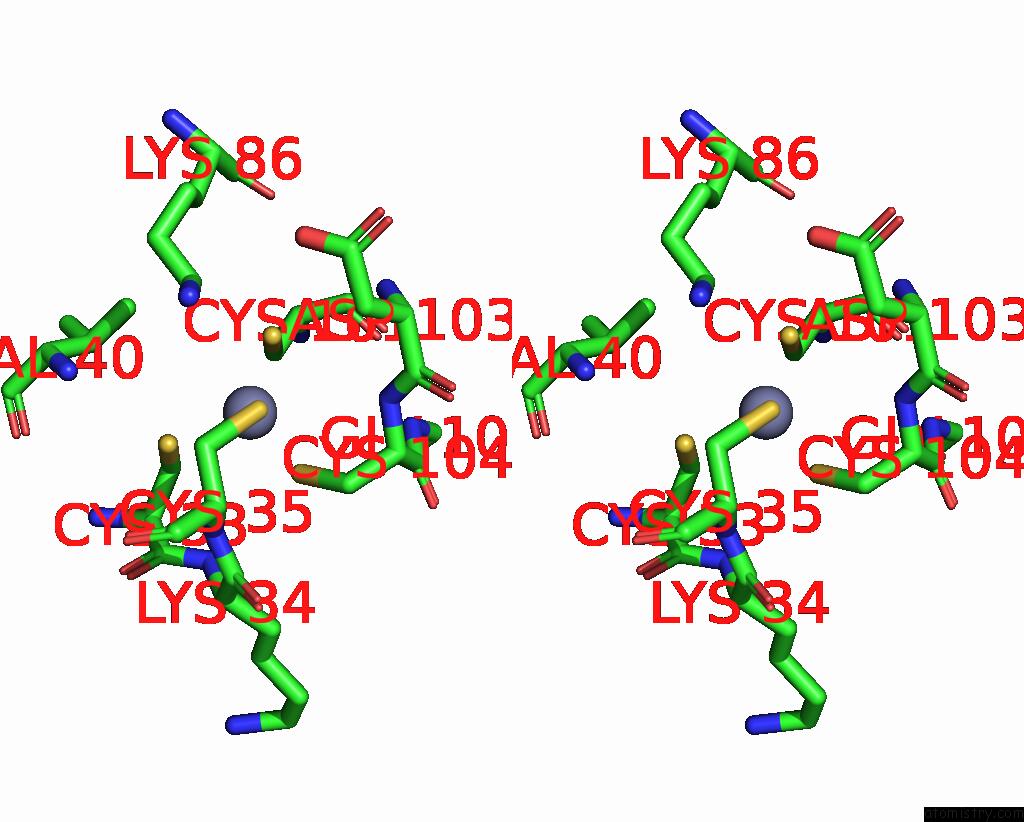
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa) within 5.0Å range:
|
Zinc binding site 4 out of 8 in 1x6m
Go back to
Zinc binding site 4 out
of 8 in the Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa)
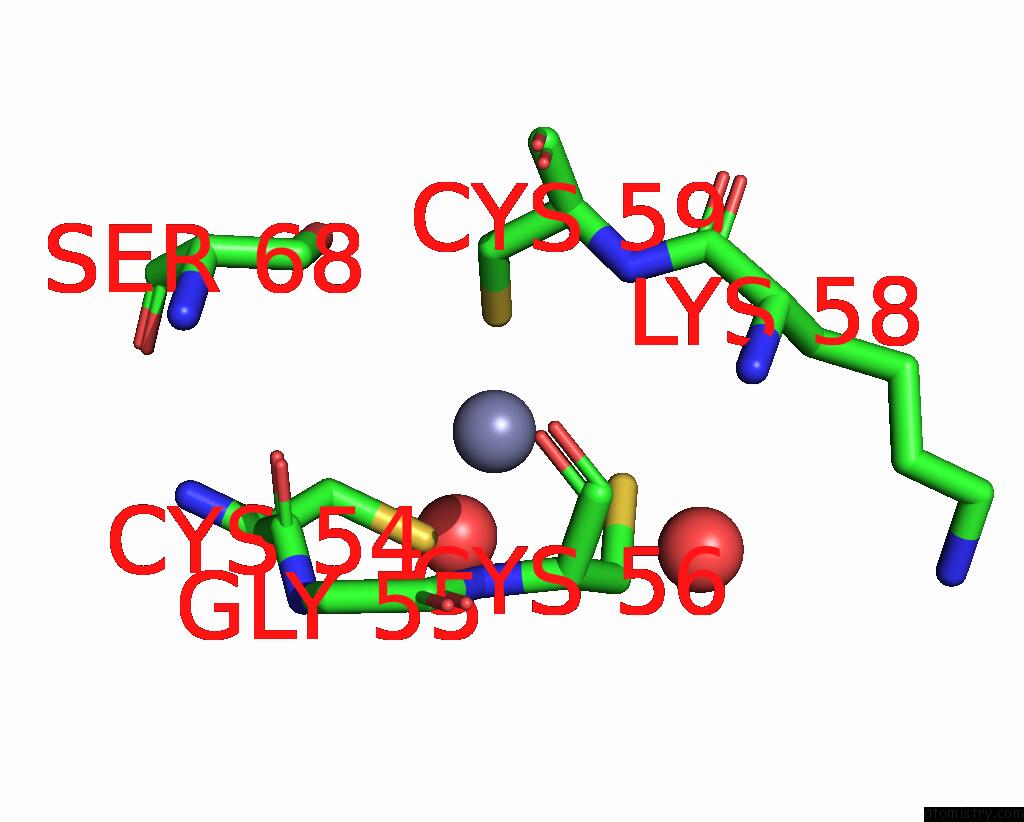
Mono view
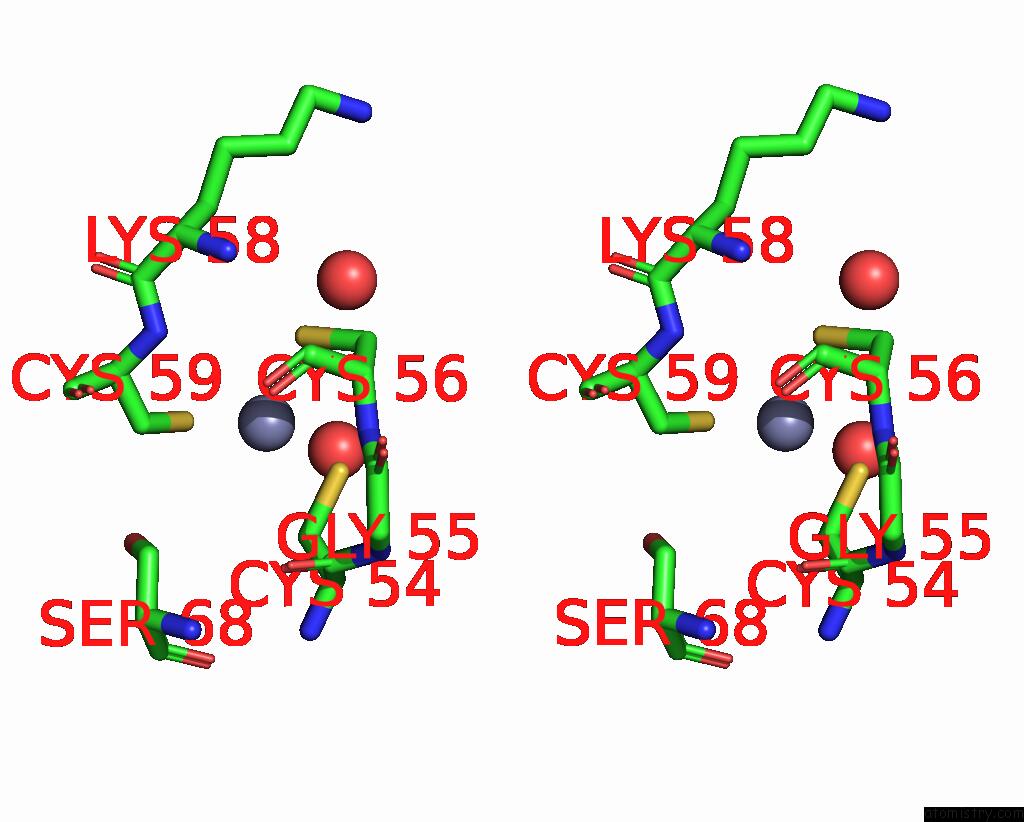
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa) within 5.0Å range:
|
Zinc binding site 5 out of 8 in 1x6m
Go back to
Zinc binding site 5 out
of 8 in the Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa)
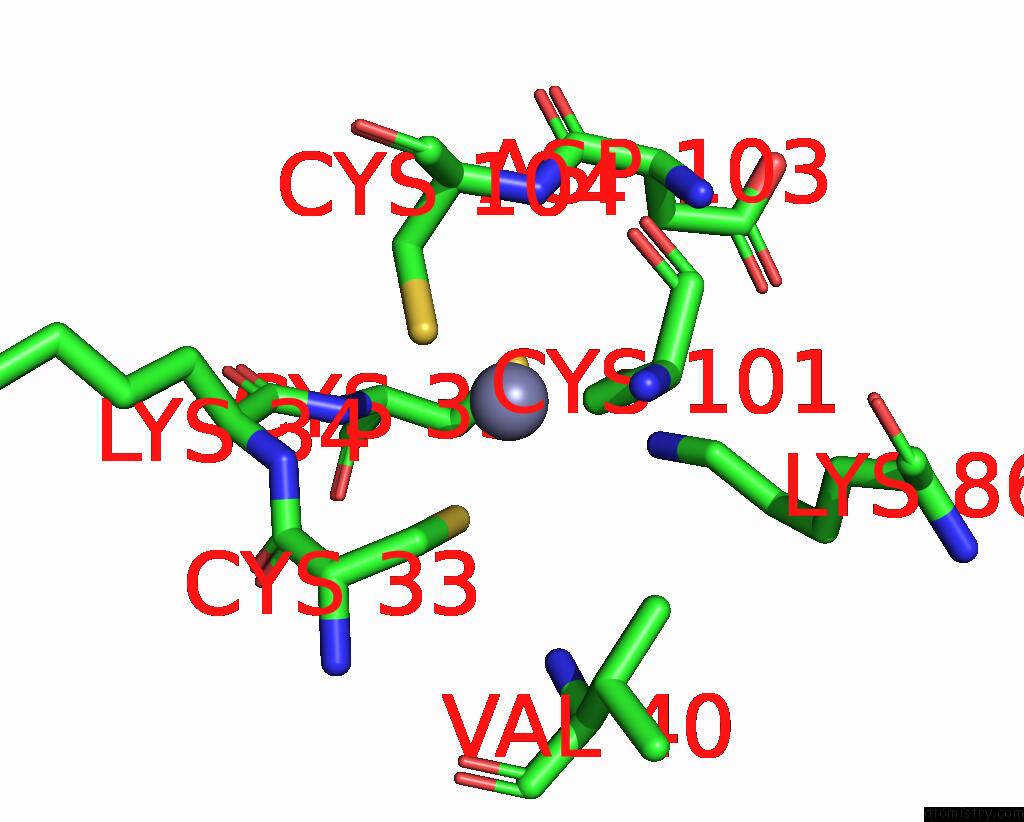
Mono view
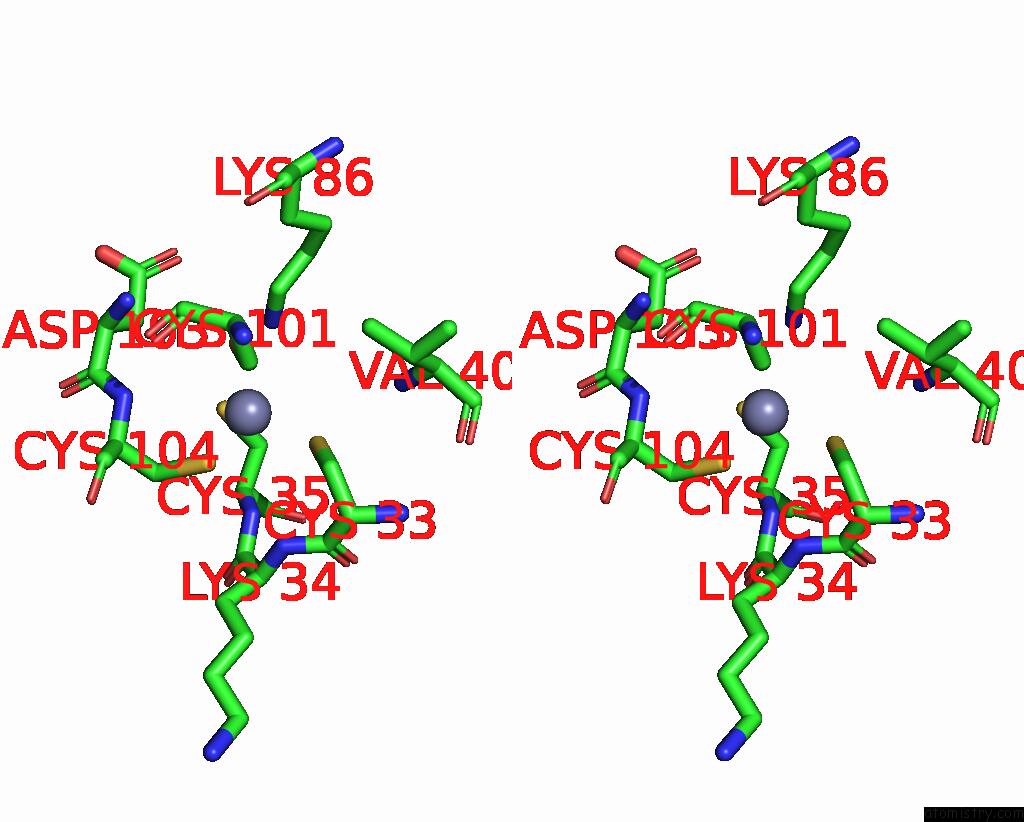
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa) within 5.0Å range:
|
Zinc binding site 6 out of 8 in 1x6m
Go back to
Zinc binding site 6 out
of 8 in the Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa)
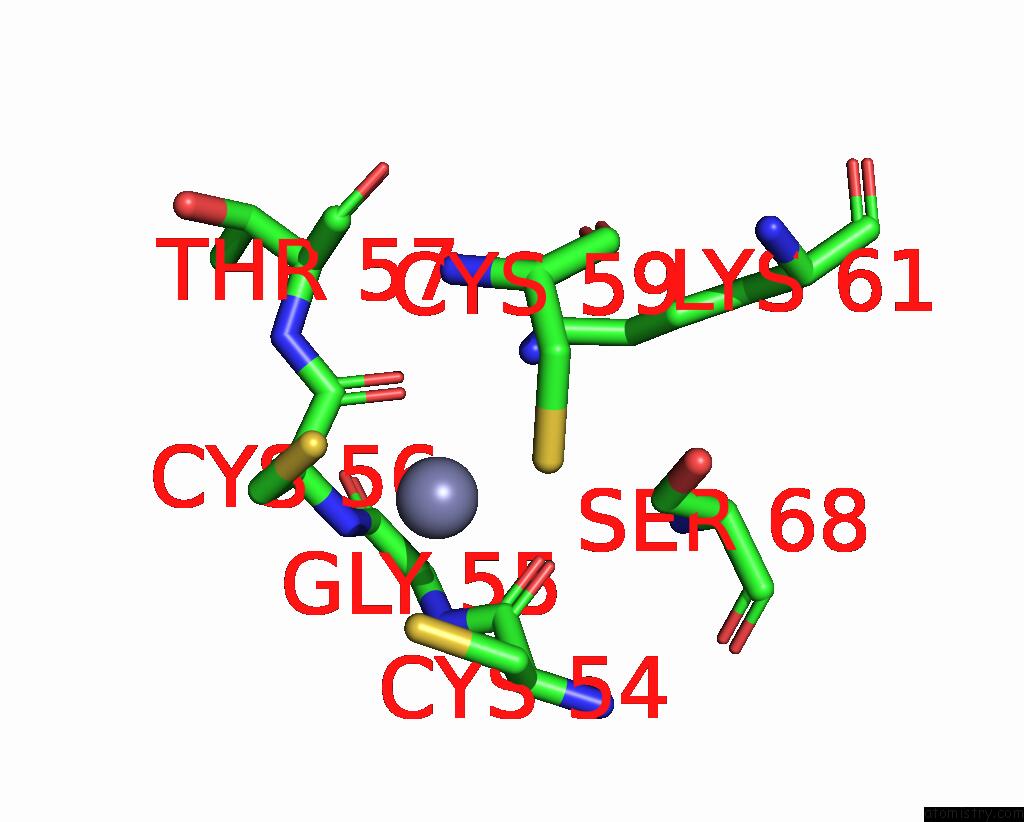
Mono view
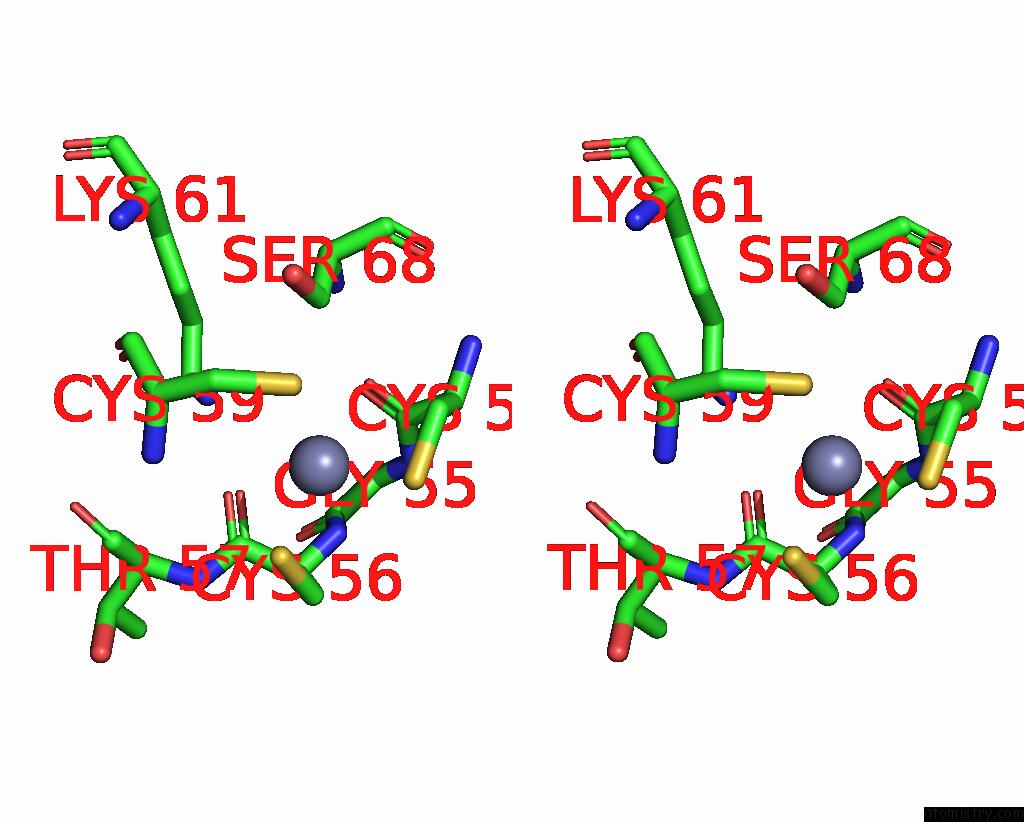
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa) within 5.0Å range:
|
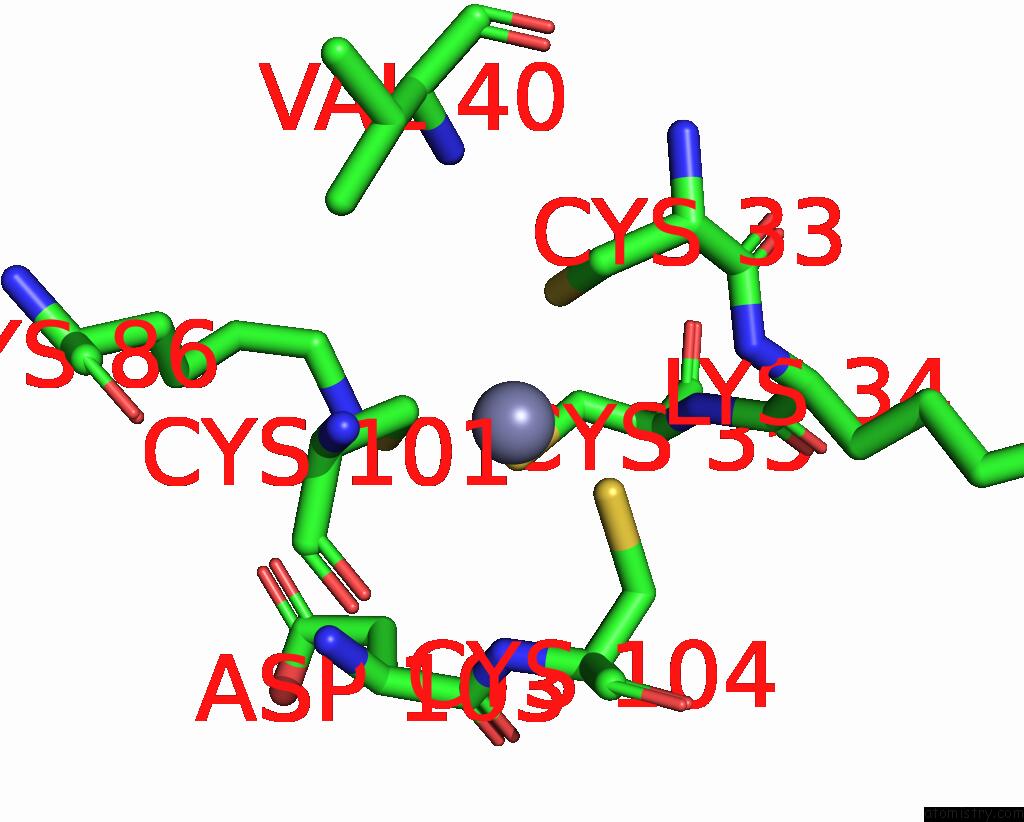
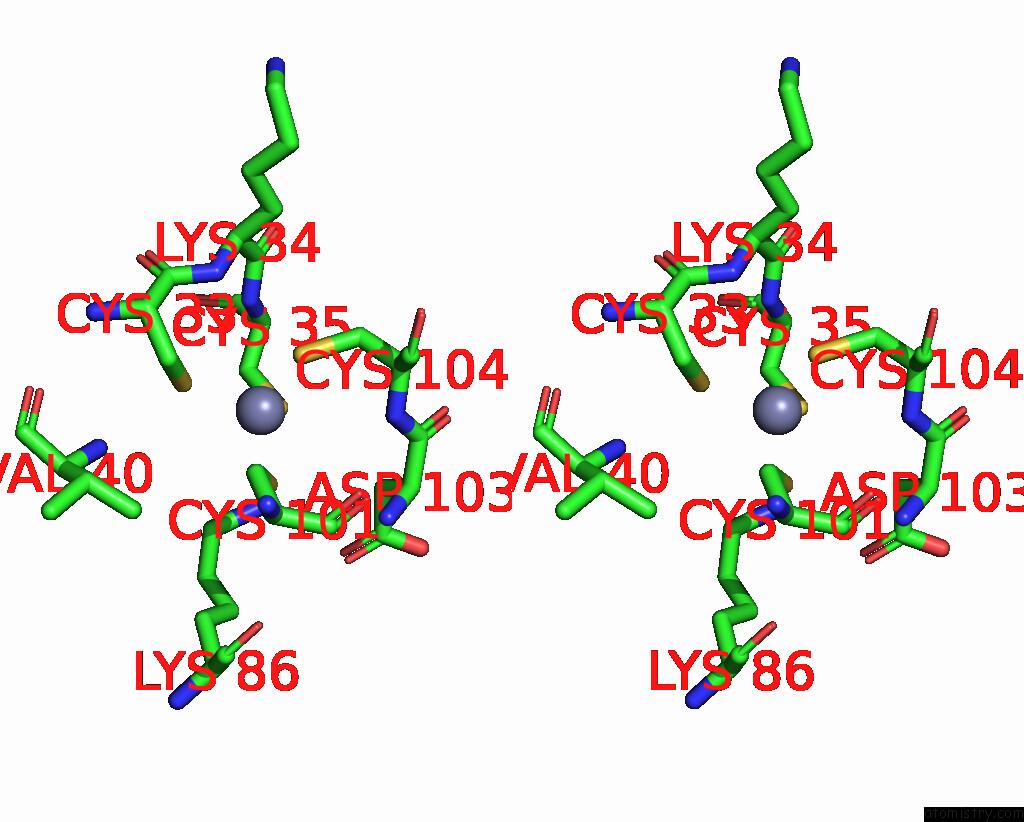
Zinc binding site 7 out of 8 in 1x6m
Go back to
Zinc binding site 7 out
of 8 in the Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa) within 5.0Å range:
|
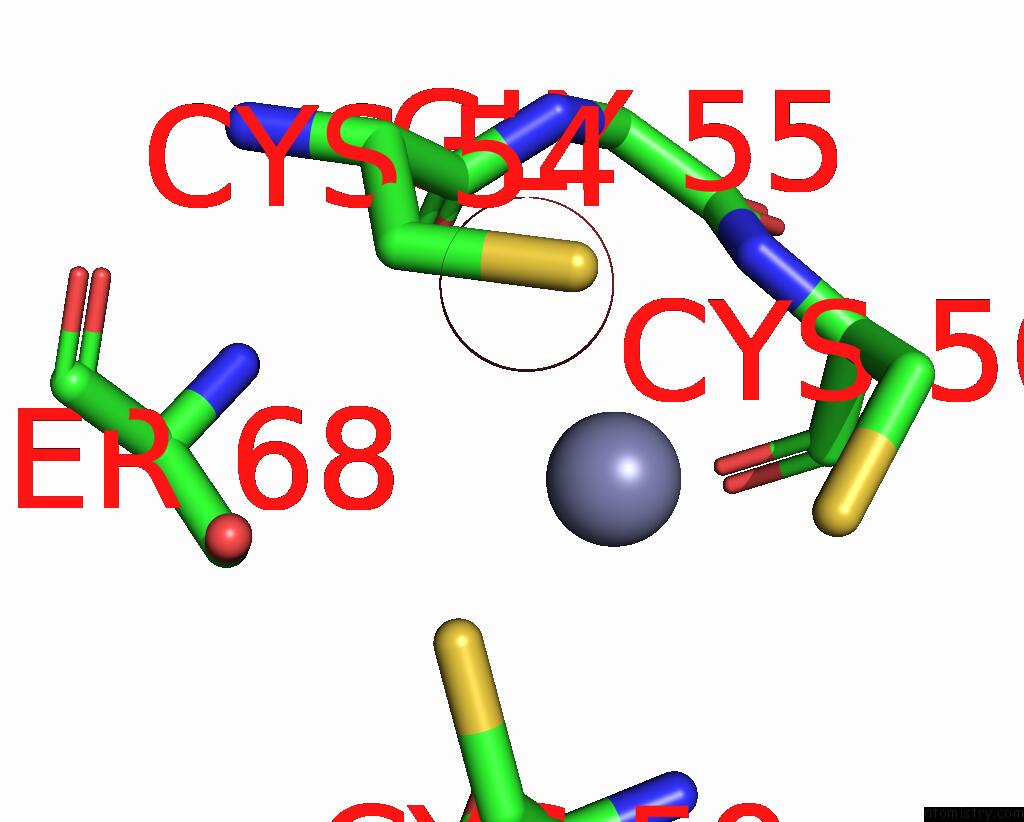
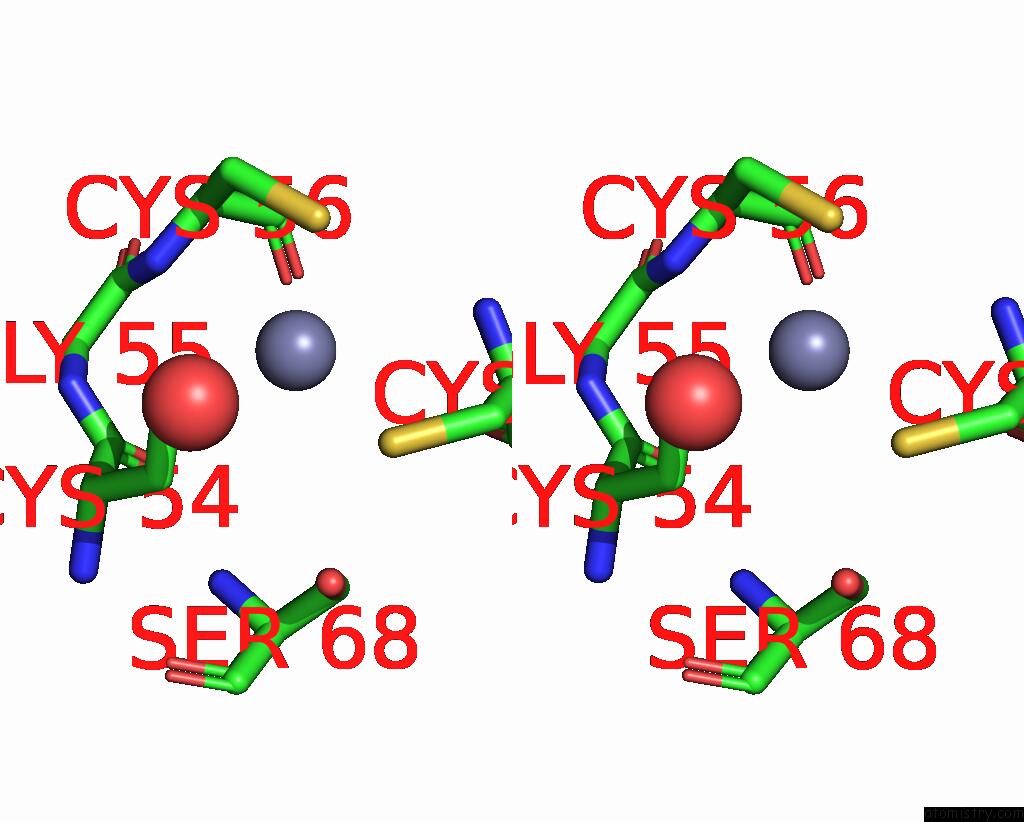
Zinc binding site 8 out of 8 in 1x6m
Go back to
Zinc binding site 8 out
of 8 in the Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 8 of Crystal Structure of the Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa) within 5.0Å range:
|
Reference:
A.M.Neculai,
D.Neculai,
C.Griesinger,
J.A.Vorholt,
S.Becker.
A Dynamic Zinc Redox Switch J.Biol.Chem. V. 280 2826 2005.
ISSN: ISSN 0021-9258
PubMed: 15548539
DOI: 10.1074/JBC.C400517200
Page generated: Wed Aug 20 00:12:39 2025
ISSN: ISSN 0021-9258
PubMed: 15548539
DOI: 10.1074/JBC.C400517200
Last articles
Zn in 2GZGZn in 2GZF
Zn in 2GZE
Zn in 2GVI
Zn in 2GYK
Zn in 2GWN
Zn in 2GWG
Zn in 2GVF
Zn in 2GVM
Zn in 2GU2