Zinc »
PDB 1tbt-1to4 »
1tjl »
Zinc in PDB 1tjl: Crystal Structure of Transcription Factor Dksa From E. Coli
Protein crystallography data
The structure of Crystal Structure of Transcription Factor Dksa From E. Coli, PDB code: 1tjl
was solved by
A.Perederina,
V.Svetlov,
M.N.Vassylyeva,
I.Artsimovitch,
S.Yokoyama,
D.G.Vassylyev,
Riken Structural Genomics/Proteomics Initiative (Rsgi),
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 38.95 / 2.00 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 91.319, 96.593, 117.477, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 22.8 / 26.8 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Crystal Structure of Transcription Factor Dksa From E. Coli
(pdb code 1tjl). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 10 binding sites of Zinc where determined in the Crystal Structure of Transcription Factor Dksa From E. Coli, PDB code: 1tjl:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
In total 10 binding sites of Zinc where determined in the Crystal Structure of Transcription Factor Dksa From E. Coli, PDB code: 1tjl:
Jump to Zinc binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Zinc binding site 1 out of 10 in 1tjl
Go back to
Zinc binding site 1 out
of 10 in the Crystal Structure of Transcription Factor Dksa From E. Coli
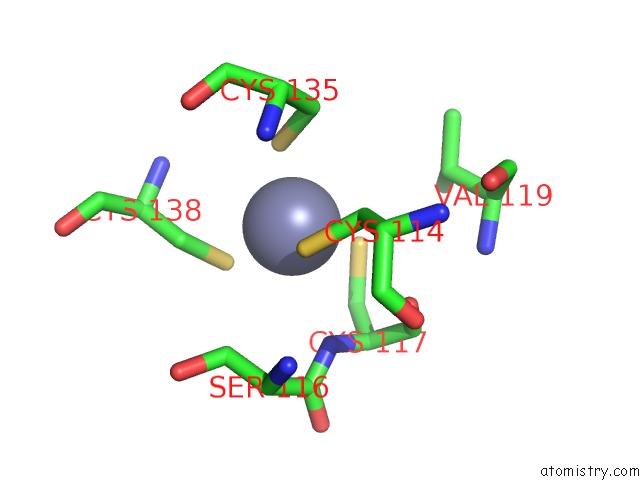
Mono view
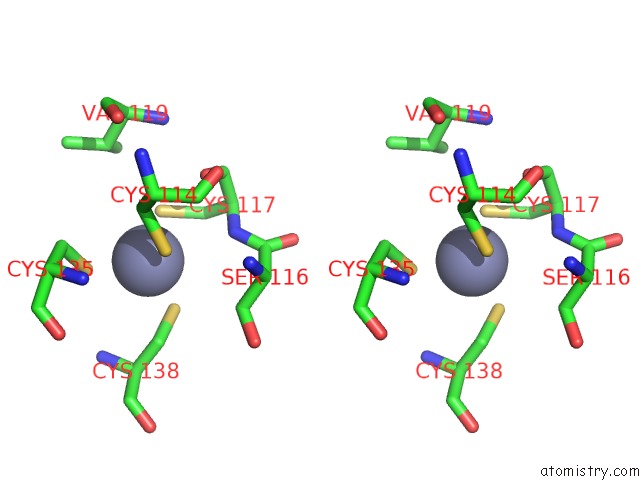
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Crystal Structure of Transcription Factor Dksa From E. Coli within 5.0Å range:
|
Zinc binding site 2 out of 10 in 1tjl
Go back to
Zinc binding site 2 out
of 10 in the Crystal Structure of Transcription Factor Dksa From E. Coli
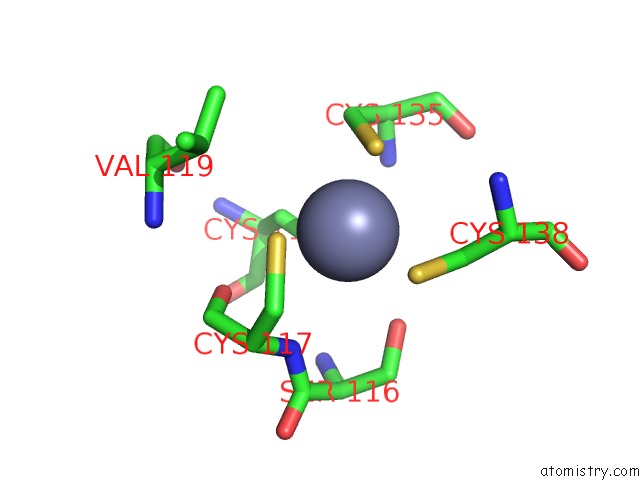
Mono view
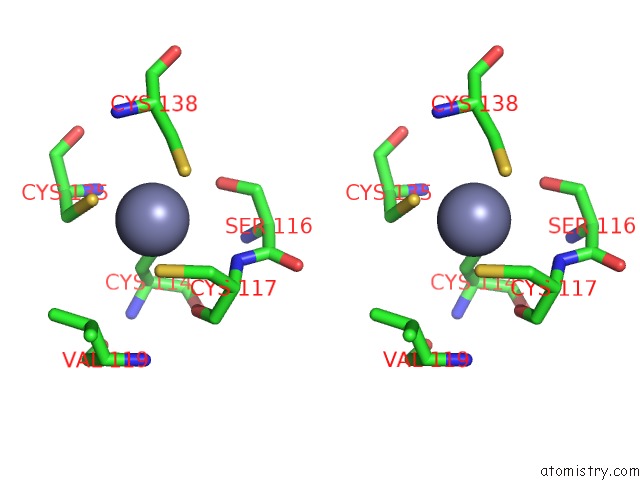
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Crystal Structure of Transcription Factor Dksa From E. Coli within 5.0Å range:
|
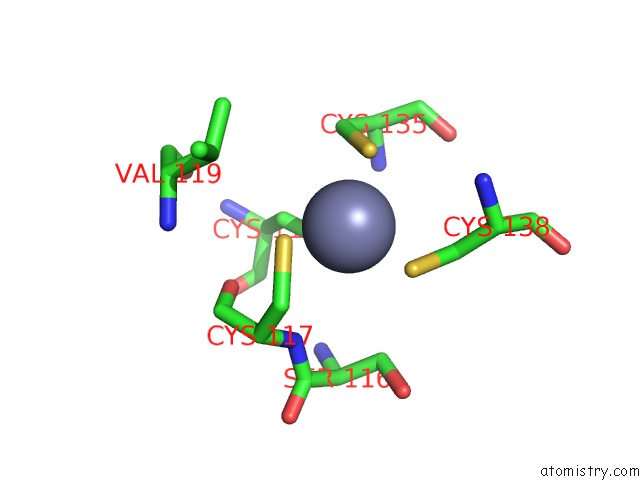
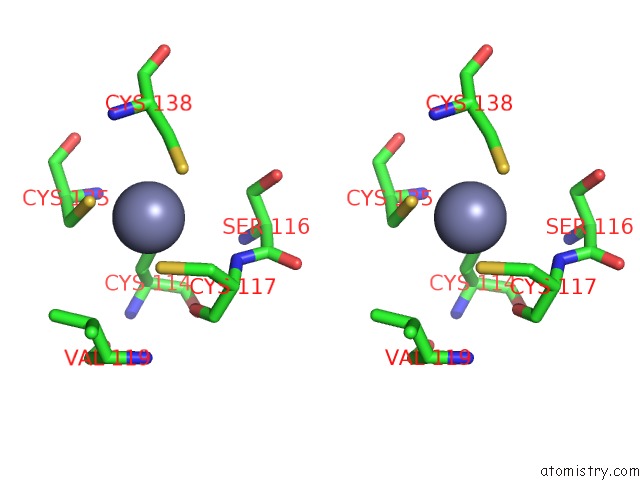
Zinc binding site 3 out of 10 in 1tjl
Go back to
Zinc binding site 3 out
of 10 in the Crystal Structure of Transcription Factor Dksa From E. Coli
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Crystal Structure of Transcription Factor Dksa From E. Coli within 5.0Å range:
|
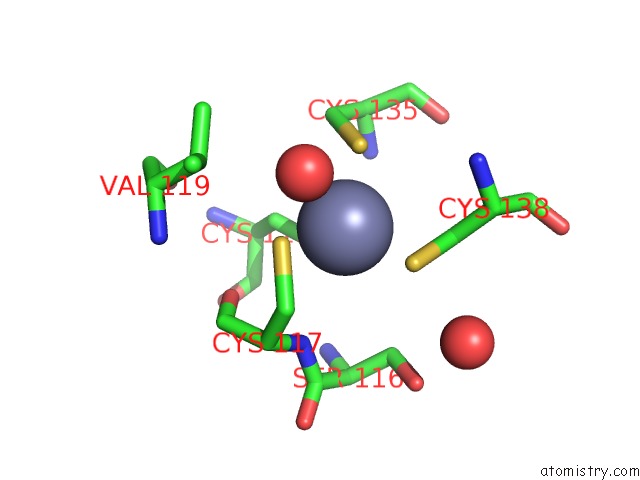
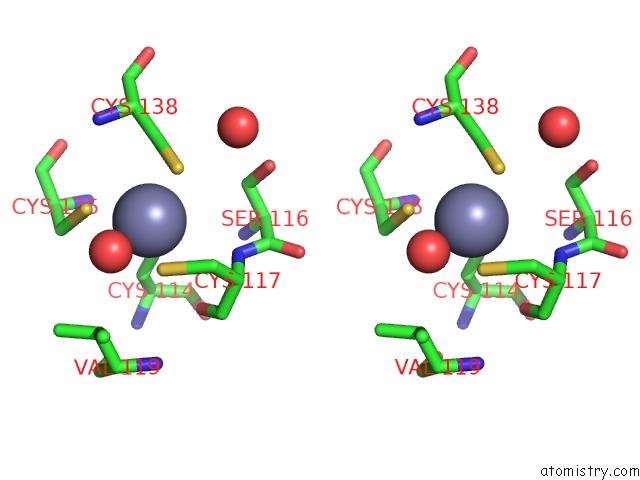
Zinc binding site 4 out of 10 in 1tjl
Go back to
Zinc binding site 4 out
of 10 in the Crystal Structure of Transcription Factor Dksa From E. Coli
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Crystal Structure of Transcription Factor Dksa From E. Coli within 5.0Å range:
|
Zinc binding site 5 out of 10 in 1tjl
Go back to
Zinc binding site 5 out
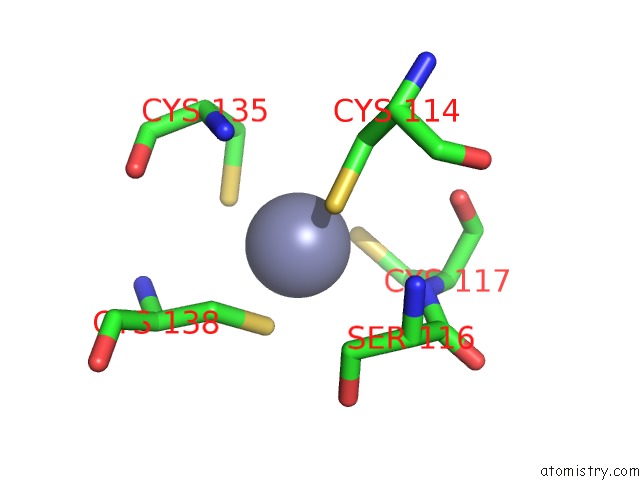
of 10 in the Crystal Structure of Transcription Factor Dksa From E. Coli
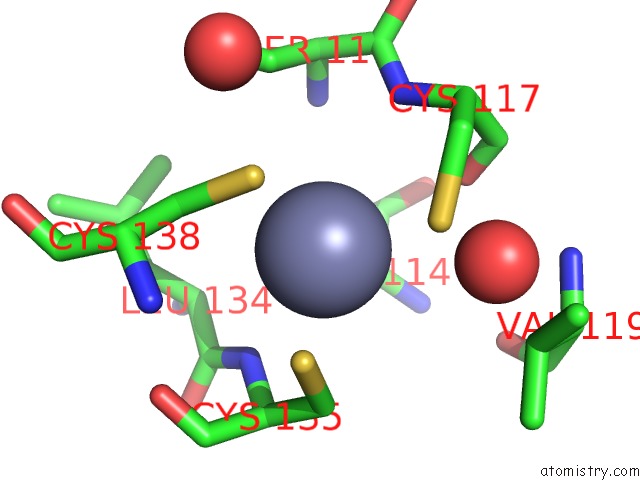
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Crystal Structure of Transcription Factor Dksa From E. Coli within 5.0Å range:
|
Zinc binding site 6 out of 10 in 1tjl
Go back to
Zinc binding site 6 out
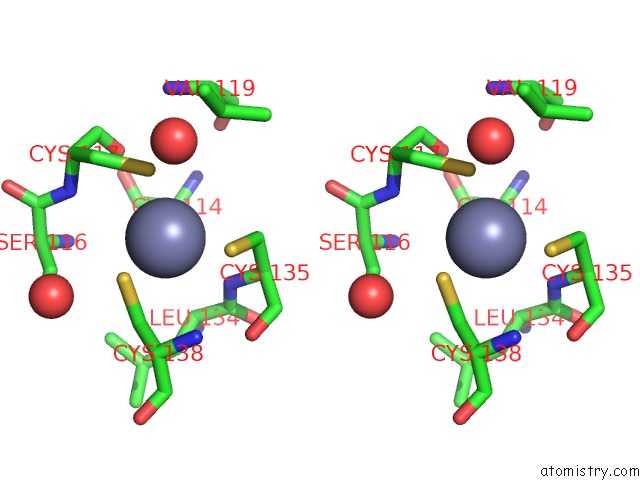
of 10 in the Crystal Structure of Transcription Factor Dksa From E. Coli
Mono view
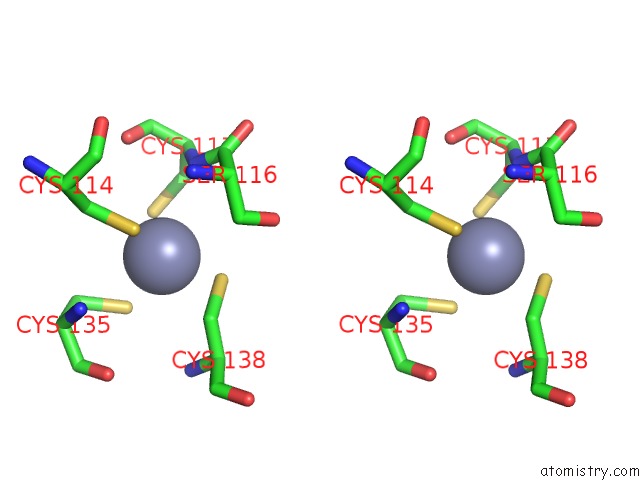
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 6 of Crystal Structure of Transcription Factor Dksa From E. Coli within 5.0Å range:
|
Zinc binding site 7 out of 10 in 1tjl
Go back to
Zinc binding site 7 out
of 10 in the Crystal Structure of Transcription Factor Dksa From E. Coli
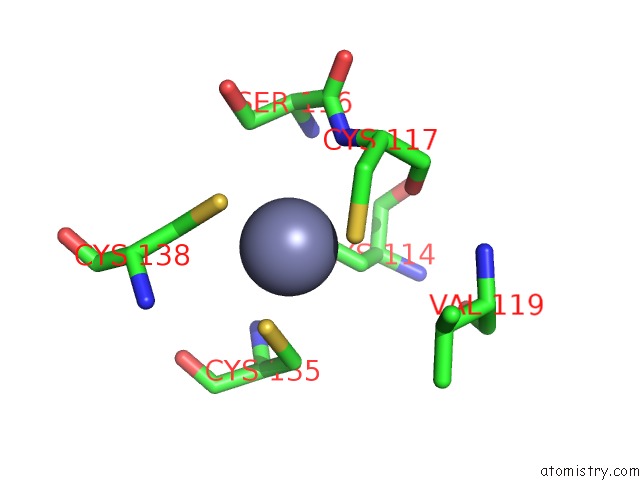
Mono view
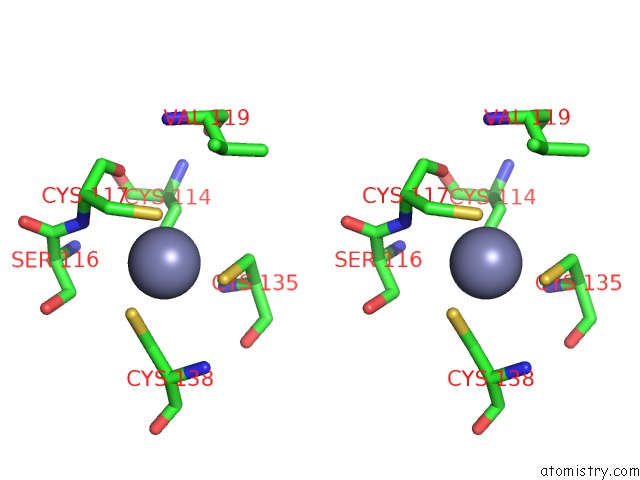
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 7 of Crystal Structure of Transcription Factor Dksa From E. Coli within 5.0Å range:
|
Zinc binding site 8 out of 10 in 1tjl
Go back to
Zinc binding site 8 out
of 10 in the Crystal Structure of Transcription Factor Dksa From E. Coli
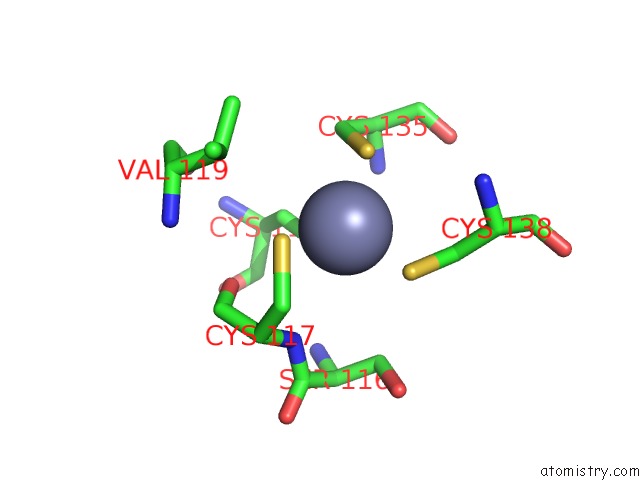
Mono view
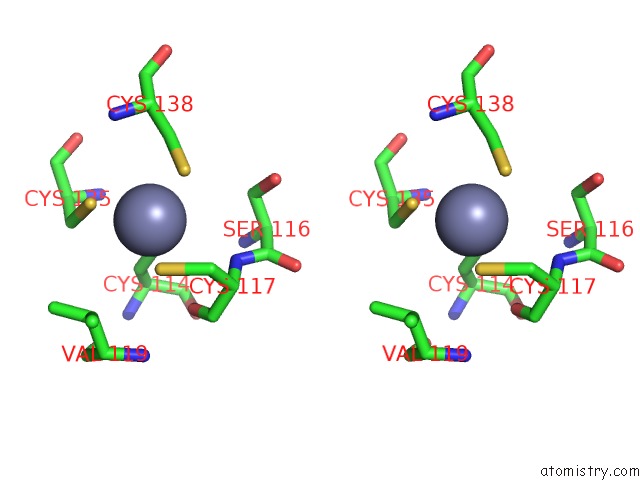
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 8 of Crystal Structure of Transcription Factor Dksa From E. Coli within 5.0Å range:
|
Zinc binding site 9 out of 10 in 1tjl
Go back to
Zinc binding site 9 out
of 10 in the Crystal Structure of Transcription Factor Dksa From E. Coli
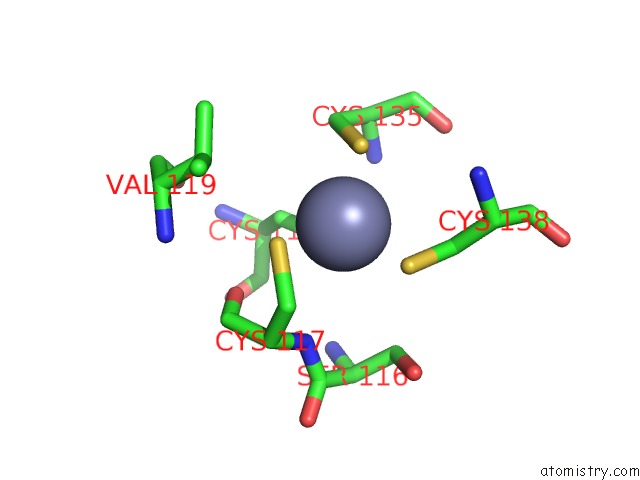
Mono view
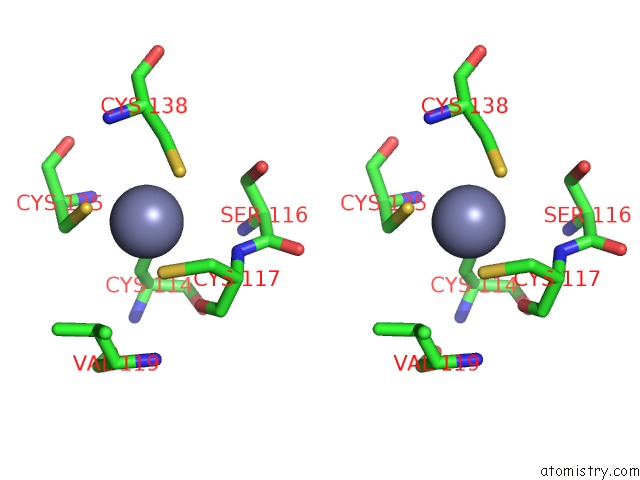
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 9 of Crystal Structure of Transcription Factor Dksa From E. Coli within 5.0Å range:
|
Zinc binding site 10 out of 10 in 1tjl
Go back to
Zinc binding site 10 out
of 10 in the Crystal Structure of Transcription Factor Dksa From E. Coli
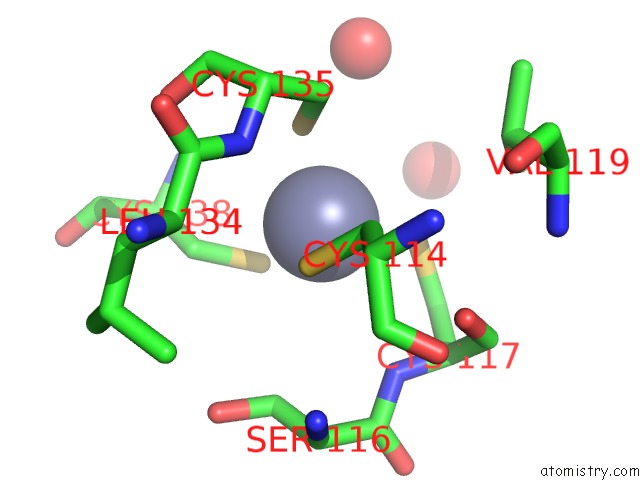
Mono view
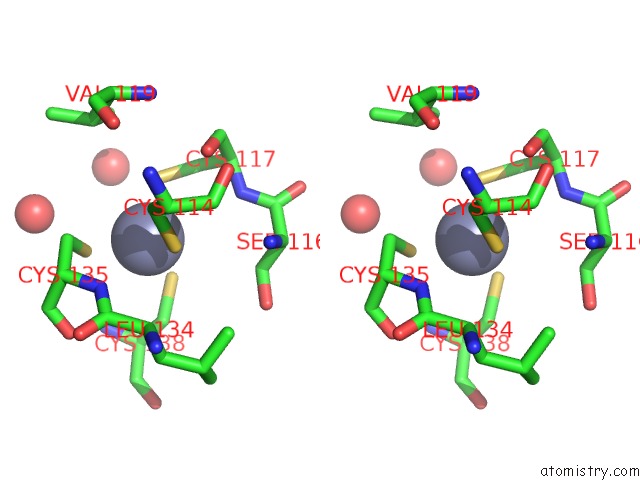
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 10 of Crystal Structure of Transcription Factor Dksa From E. Coli within 5.0Å range:
|
Reference:
A.Perederina,
V.Svetlov,
M.N.Vassylyeva,
T.H.Tahirov,
S.Yokoyama,
I.Artsimovitch,
D.G.Vassylyev.
Regulation Through the Secondary Channel--Structural Framework For Ppgpp-Dksa Synergism During Transcription Cell(Cambridge,Mass.) V. 118 297 2004.
ISSN: ISSN 0092-8674
PubMed: 15294156
DOI: 10.1016/J.CELL.2004.06.030
Page generated: Wed Oct 16 19:09:08 2024
ISSN: ISSN 0092-8674
PubMed: 15294156
DOI: 10.1016/J.CELL.2004.06.030
Last articles
As in 4B09As in 4CFT
As in 4CAR
As in 4BKT
As in 4C3A
As in 4C2A
As in 4AWJ
As in 4BKS
As in 3WGH
As in 3ZPG