Zinc »
PDB 1tbt-1to4 »
1tfi »
Zinc in PDB 1tfi: A Novel Zn Finger Motif in the Basal Transcriptional Machinery: Three-Dimensional uc(Nmr) Studies of the Nucleic- Acid Binding Domain of Transcriptional Elongation Factor Tfiis
Zinc Binding Sites:
The binding sites of Zinc atom in the A Novel Zn Finger Motif in the Basal Transcriptional Machinery: Three-Dimensional uc(Nmr) Studies of the Nucleic- Acid Binding Domain of Transcriptional Elongation Factor Tfiis
(pdb code 1tfi). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the A Novel Zn Finger Motif in the Basal Transcriptional Machinery: Three-Dimensional uc(Nmr) Studies of the Nucleic- Acid Binding Domain of Transcriptional Elongation Factor Tfiis, PDB code: 1tfi:
In total only one binding site of Zinc was determined in the A Novel Zn Finger Motif in the Basal Transcriptional Machinery: Three-Dimensional uc(Nmr) Studies of the Nucleic- Acid Binding Domain of Transcriptional Elongation Factor Tfiis, PDB code: 1tfi:
Zinc binding site 1 out of 1 in 1tfi
Go back to
Zinc binding site 1 out
of 1 in the A Novel Zn Finger Motif in the Basal Transcriptional Machinery: Three-Dimensional uc(Nmr) Studies of the Nucleic- Acid Binding Domain of Transcriptional Elongation Factor Tfiis
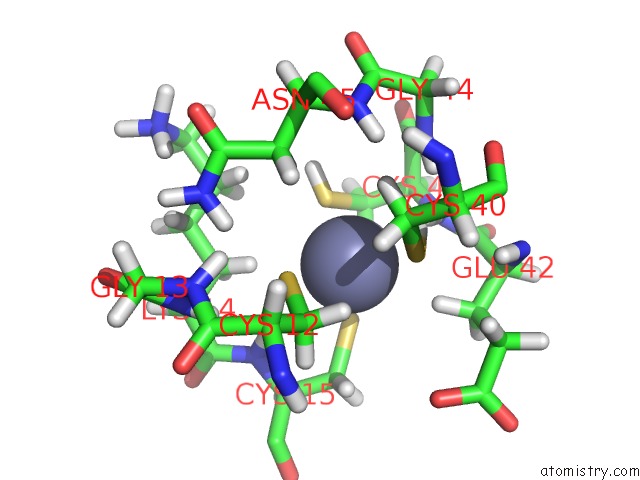
Mono view
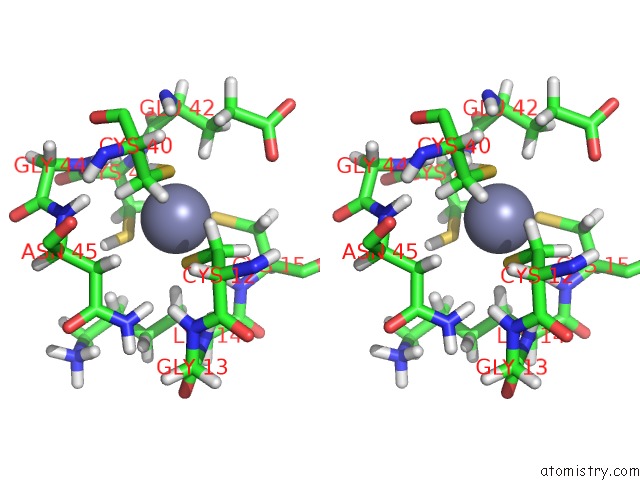
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of A Novel Zn Finger Motif in the Basal Transcriptional Machinery: Three-Dimensional uc(Nmr) Studies of the Nucleic- Acid Binding Domain of Transcriptional Elongation Factor Tfiis within 5.0Å range:
|
Reference:
X.Qian,
S.N.Gozani,
H.Yoon,
C.J.Jeon,
K.Agarwal,
M.A.Weiss.
Novel Zinc Finger Motif in the Basal Transcriptional Machinery: Three-Dimensional uc(Nmr) Studies of the Nucleic Acid Binding Domain of Transcriptional Elongation Factor Tfiis. Biochemistry V. 32 9944 1993.
ISSN: ISSN 0006-2960
PubMed: 8399164
DOI: 10.1021/BI00089A010
Page generated: Tue Aug 19 23:20:22 2025
ISSN: ISSN 0006-2960
PubMed: 8399164
DOI: 10.1021/BI00089A010
Last articles
Zn in 2ANUZn in 2AQR
Zn in 2AQQ
Zn in 2AQO
Zn in 2AQP
Zn in 2AQN
Zn in 2APS
Zn in 2AQC
Zn in 2AQ2
Zn in 2APO