Zinc »
PDB 1lgd-1m2n »
1lv3 »
Zinc in PDB 1lv3: Solution uc(Nmr) Structure of Zinc Finger Protein Yacg From Escherichia Coli. Northeast Structural Genomics Consortium Target ET92.
Zinc Binding Sites:
The binding sites of Zinc atom in the Solution uc(Nmr) Structure of Zinc Finger Protein Yacg From Escherichia Coli. Northeast Structural Genomics Consortium Target ET92.
(pdb code 1lv3). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Solution uc(Nmr) Structure of Zinc Finger Protein Yacg From Escherichia Coli. Northeast Structural Genomics Consortium Target ET92., PDB code: 1lv3:
In total only one binding site of Zinc was determined in the Solution uc(Nmr) Structure of Zinc Finger Protein Yacg From Escherichia Coli. Northeast Structural Genomics Consortium Target ET92., PDB code: 1lv3:
Zinc binding site 1 out of 1 in 1lv3
Go back to
Zinc binding site 1 out
of 1 in the Solution uc(Nmr) Structure of Zinc Finger Protein Yacg From Escherichia Coli. Northeast Structural Genomics Consortium Target ET92.
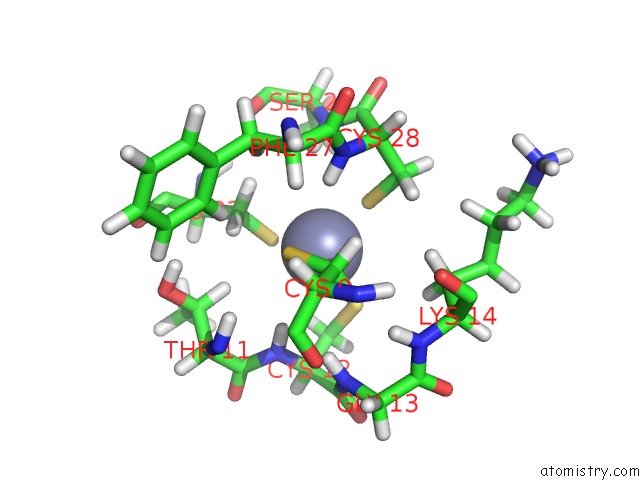
Mono view
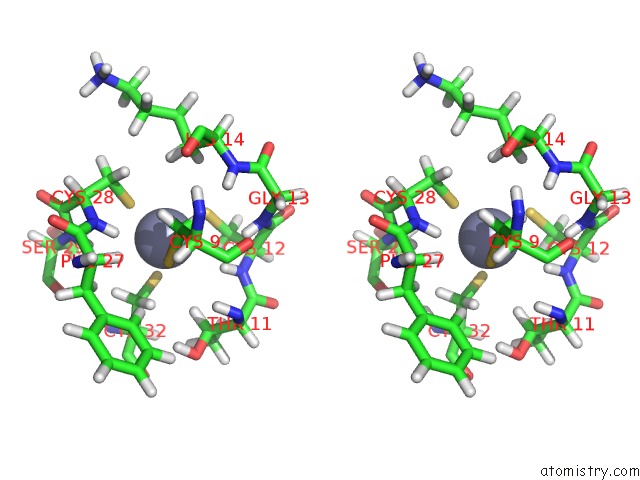
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution uc(Nmr) Structure of Zinc Finger Protein Yacg From Escherichia Coli. Northeast Structural Genomics Consortium Target ET92. within 5.0Å range:
|
Reference:
T.A.Ramelot,
J.R.Cort,
A.A.Yee,
A.Semesi,
A.M.Edwards,
C.H.Arrowsmith,
M.A.Kennedy.
uc(Nmr) Structure of the Escherichia Coli Protein Yacg: A Novel Sequence Motif in the Zinc-Finger Family of Proteins. Proteins V. 49 289 2002.
ISSN: ISSN 0887-3585
PubMed: 12211008
DOI: 10.1002/PROT.10214
Page generated: Sun Oct 13 05:14:31 2024
ISSN: ISSN 0887-3585
PubMed: 12211008
DOI: 10.1002/PROT.10214
Last articles
As in 1WMBAs in 1VHD
As in 1WCK
As in 1VHO
As in 1W2K
As in 1W0Y
As in 1TAD
As in 1TND
As in 1TYE
As in 1TZA