Zinc »
PDB 1kzy-1lg6 »
1l2c »
Zinc in PDB 1l2c: Mutm (Fpg)-Dna Estranged Thymine Mismatch Recognition Complex
Protein crystallography data
The structure of Mutm (Fpg)-Dna Estranged Thymine Mismatch Recognition Complex, PDB code: 1l2c
was solved by
J.C.Fromme,
G.L.Verdine,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 50.00 / 2.20 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 45.219, 95.005, 103.552, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21.3 / 24.7 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Mutm (Fpg)-Dna Estranged Thymine Mismatch Recognition Complex
(pdb code 1l2c). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Mutm (Fpg)-Dna Estranged Thymine Mismatch Recognition Complex, PDB code: 1l2c:
In total only one binding site of Zinc was determined in the Mutm (Fpg)-Dna Estranged Thymine Mismatch Recognition Complex, PDB code: 1l2c:
Zinc binding site 1 out of 1 in 1l2c
Go back to
Zinc binding site 1 out
of 1 in the Mutm (Fpg)-Dna Estranged Thymine Mismatch Recognition Complex
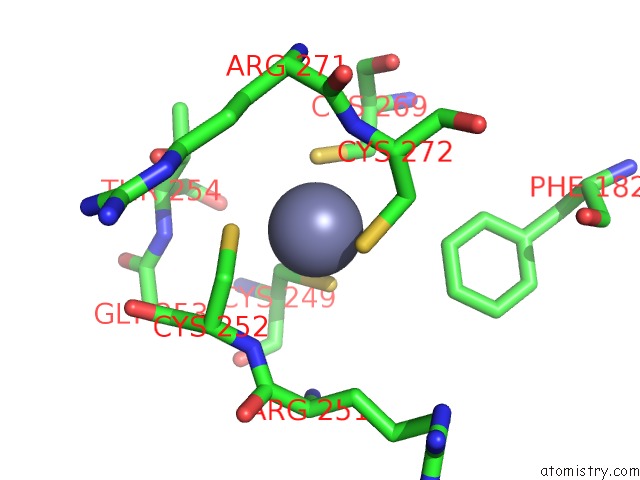
Mono view
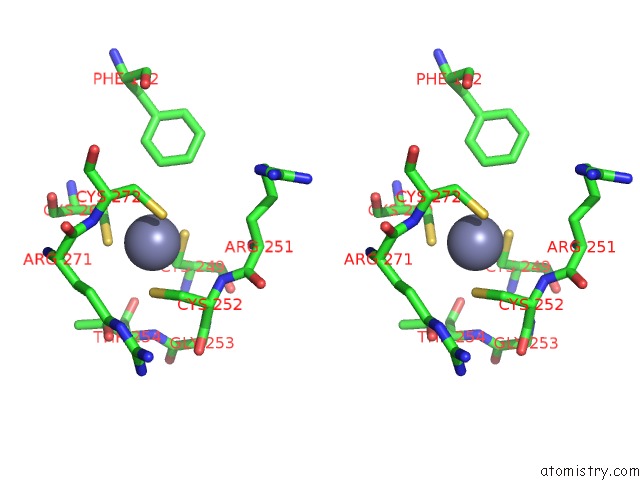
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Mutm (Fpg)-Dna Estranged Thymine Mismatch Recognition Complex within 5.0Å range:
|
Reference:
J.C.Fromme,
G.L.Verdine.
Structural Insights Into Lesion Recognition and Repair By the Bacterial 8-Oxoguanine Dna Glycosylase Mutm. Nat.Struct.Biol. V. 9 544 2002.
ISSN: ISSN 1072-8368
PubMed: 12055620
Page generated: Tue Aug 19 21:27:31 2025
ISSN: ISSN 1072-8368
PubMed: 12055620
Last articles
Zn in 1WJVZn in 1WKF
Zn in 1WKE
Zn in 1WKD
Zn in 1WJD
Zn in 1WJB
Zn in 1WJC
Zn in 1WJA
Zn in 1WJ2
Zn in 1WIM