Zinc »
PDB 1hlk-1hxr »
1hvn »
Zinc in PDB 1hvn: Zinc-and Sequence-Dependent Binding to Nucleic Acids By the N-Terminal Zinc Finger Domain of the Hiv-1 Nucleocapsid Protein: uc(Nmr) Structure of the Complex with the Psi-Site Analog, D/Acgcc
Zinc Binding Sites:
The binding sites of Zinc atom in the Zinc-and Sequence-Dependent Binding to Nucleic Acids By the N-Terminal Zinc Finger Domain of the Hiv-1 Nucleocapsid Protein: uc(Nmr) Structure of the Complex with the Psi-Site Analog, D/Acgcc
(pdb code 1hvn). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total only one binding site of Zinc was determined in the Zinc-and Sequence-Dependent Binding to Nucleic Acids By the N-Terminal Zinc Finger Domain of the Hiv-1 Nucleocapsid Protein: uc(Nmr) Structure of the Complex with the Psi-Site Analog, D/Acgcc, PDB code: 1hvn:
In total only one binding site of Zinc was determined in the Zinc-and Sequence-Dependent Binding to Nucleic Acids By the N-Terminal Zinc Finger Domain of the Hiv-1 Nucleocapsid Protein: uc(Nmr) Structure of the Complex with the Psi-Site Analog, D/Acgcc, PDB code: 1hvn:
Zinc binding site 1 out of 1 in 1hvn
Go back to
Zinc binding site 1 out
of 1 in the Zinc-and Sequence-Dependent Binding to Nucleic Acids By the N-Terminal Zinc Finger Domain of the Hiv-1 Nucleocapsid Protein: uc(Nmr) Structure of the Complex with the Psi-Site Analog, D/Acgcc
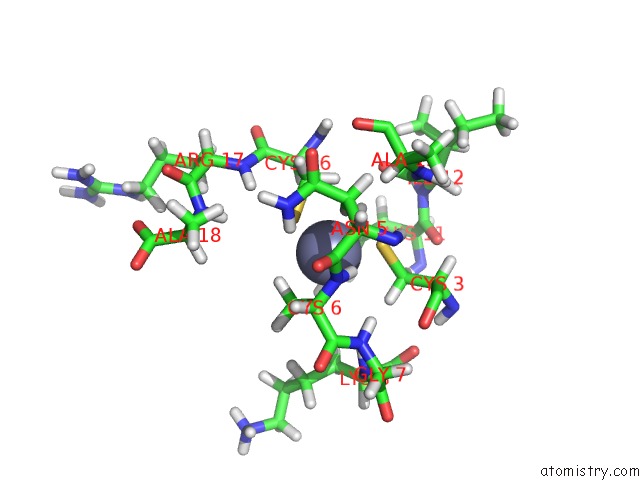
Mono view
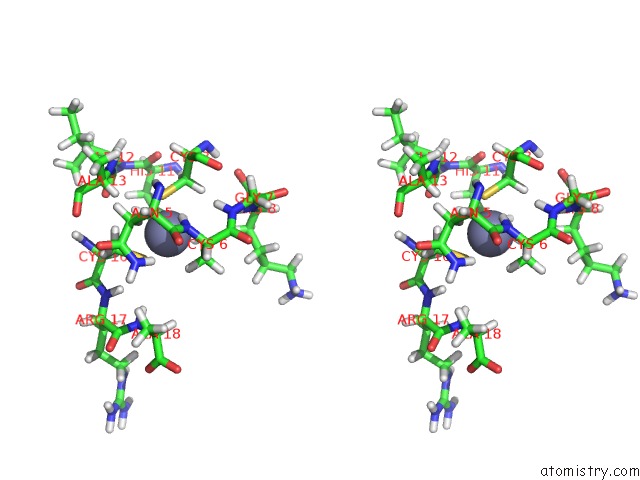
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Zinc-and Sequence-Dependent Binding to Nucleic Acids By the N-Terminal Zinc Finger Domain of the Hiv-1 Nucleocapsid Protein: uc(Nmr) Structure of the Complex with the Psi-Site Analog, D/Acgcc within 5.0Å range:
|
Reference:
T.L.South,
M.F.Summers.
Zinc- and Sequence-Dependent Binding to Nucleic Acids By the N-Terminal Zinc Finger of the Hiv-1 Nucleocapsid Protein: uc(Nmr) Structure of the Complex with the Psi-Site Analog, Dacgcc. Protein Sci. V. 2 3 1993.
ISSN: ISSN 0961-8368
PubMed: 8443588
Page generated: Tue Aug 19 20:42:49 2025
ISSN: ISSN 0961-8368
PubMed: 8443588
Last articles
Zn in 1TLXZn in 1TLP
Zn in 1TKJ
Zn in 1TLI
Zn in 1TKH
Zn in 1TF6
Zn in 1TKF
Zn in 1TIY
Zn in 1THJ
Zn in 1THL