Zinc »
PDB 1gkr-1h4t »
1h4s »
Zinc in PDB 1h4s: Prolyl-Trna Synthetase From Thermus Thermophilus Complexed with Trnapro(Cgg) and A Prolyl-Adenylate Analogue
Enzymatic activity of Prolyl-Trna Synthetase From Thermus Thermophilus Complexed with Trnapro(Cgg) and A Prolyl-Adenylate Analogue
All present enzymatic activity of Prolyl-Trna Synthetase From Thermus Thermophilus Complexed with Trnapro(Cgg) and A Prolyl-Adenylate Analogue:
6.1.1.15;
6.1.1.15;
Protein crystallography data
The structure of Prolyl-Trna Synthetase From Thermus Thermophilus Complexed with Trnapro(Cgg) and A Prolyl-Adenylate Analogue, PDB code: 1h4s
was solved by
A.Yaremchuk,
M.Tukalo,
S.Cusack,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 20 / 2.85 |
| Space group | P 43 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 140.780, 140.780, 236.970, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 21 / 24.2 |
Zinc Binding Sites:
The binding sites of Zinc atom in the Prolyl-Trna Synthetase From Thermus Thermophilus Complexed with Trnapro(Cgg) and A Prolyl-Adenylate Analogue
(pdb code 1h4s). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Prolyl-Trna Synthetase From Thermus Thermophilus Complexed with Trnapro(Cgg) and A Prolyl-Adenylate Analogue, PDB code: 1h4s:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Prolyl-Trna Synthetase From Thermus Thermophilus Complexed with Trnapro(Cgg) and A Prolyl-Adenylate Analogue, PDB code: 1h4s:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 1h4s
Go back to
Zinc binding site 1 out
of 2 in the Prolyl-Trna Synthetase From Thermus Thermophilus Complexed with Trnapro(Cgg) and A Prolyl-Adenylate Analogue
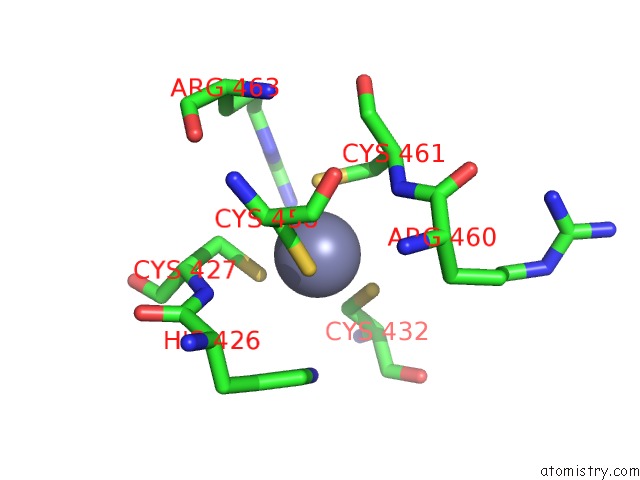
Mono view
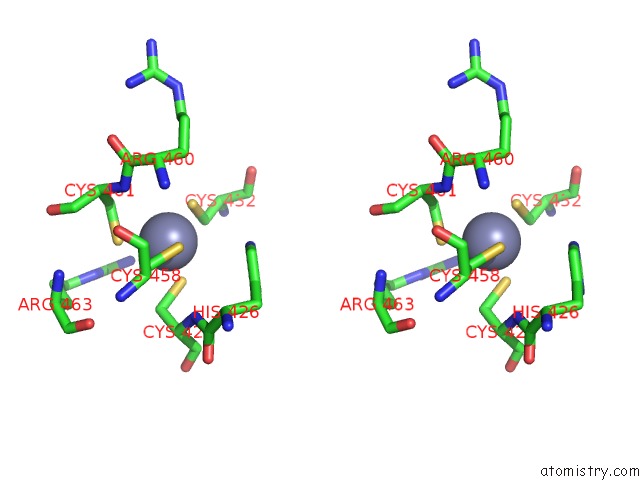
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Prolyl-Trna Synthetase From Thermus Thermophilus Complexed with Trnapro(Cgg) and A Prolyl-Adenylate Analogue within 5.0Å range:
|
Zinc binding site 2 out of 2 in 1h4s
Go back to
Zinc binding site 2 out
of 2 in the Prolyl-Trna Synthetase From Thermus Thermophilus Complexed with Trnapro(Cgg) and A Prolyl-Adenylate Analogue
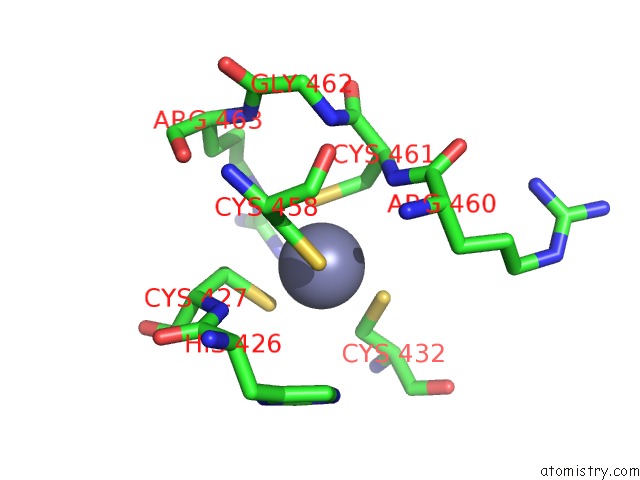
Mono view
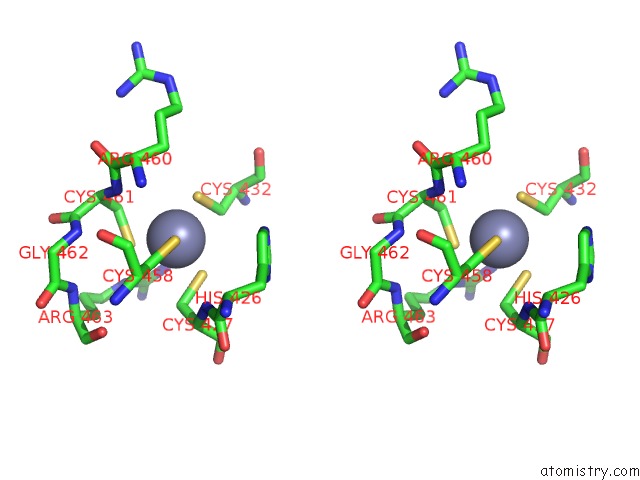
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Prolyl-Trna Synthetase From Thermus Thermophilus Complexed with Trnapro(Cgg) and A Prolyl-Adenylate Analogue within 5.0Å range:
|
Reference:
A.Yaremchuk,
M.Tukalo,
M.Grotli,
S.Cusack.
A Succession of Substrate Induced Conformational Changes Ensures the Amino Acid Specificity of Thermus Thermophilus Prolyl-Trna Synthetase: Comparison with Histidyl-Trna Synthetase J.Mol.Biol. V. 309 989 2001.
ISSN: ISSN 0022-2836
PubMed: 11399074
DOI: 10.1006/JMBI.2001.4712
Page generated: Sun Oct 13 02:00:23 2024
ISSN: ISSN 0022-2836
PubMed: 11399074
DOI: 10.1006/JMBI.2001.4712
Last articles
Al in 8SHDAl in 8SHA
Al in 8SGL
Al in 8SH9
Al in 8SGC
Al in 8SG9
Al in 8SFF
Al in 8SG8
Al in 8R1A
Al in 8Q75