Zinc »
PDB 12ca-1add »
1aaf »
Zinc in PDB 1aaf: Nucleocapsid Zinc Fingers Detected in Retroviruses: Exafs Studies on Intact Viruses and the Solution-State Structure of the Nucleocapsid Protein From Hiv-1
Zinc Binding Sites:
The binding sites of Zinc atom in the Nucleocapsid Zinc Fingers Detected in Retroviruses: Exafs Studies on Intact Viruses and the Solution-State Structure of the Nucleocapsid Protein From Hiv-1
(pdb code 1aaf). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Nucleocapsid Zinc Fingers Detected in Retroviruses: Exafs Studies on Intact Viruses and the Solution-State Structure of the Nucleocapsid Protein From Hiv-1, PDB code: 1aaf:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Nucleocapsid Zinc Fingers Detected in Retroviruses: Exafs Studies on Intact Viruses and the Solution-State Structure of the Nucleocapsid Protein From Hiv-1, PDB code: 1aaf:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 1aaf
Go back to
Zinc binding site 1 out
of 2 in the Nucleocapsid Zinc Fingers Detected in Retroviruses: Exafs Studies on Intact Viruses and the Solution-State Structure of the Nucleocapsid Protein From Hiv-1
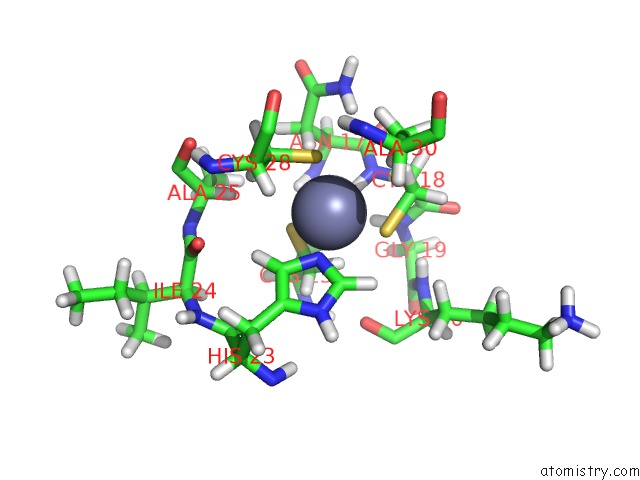
Mono view
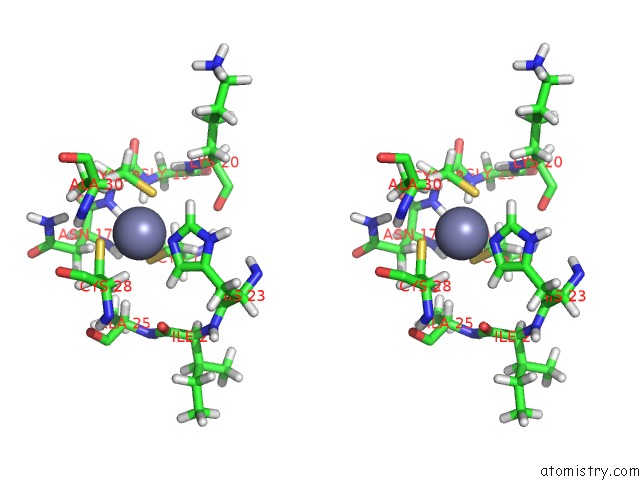
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Nucleocapsid Zinc Fingers Detected in Retroviruses: Exafs Studies on Intact Viruses and the Solution-State Structure of the Nucleocapsid Protein From Hiv-1 within 5.0Å range:
|
Zinc binding site 2 out of 2 in 1aaf
Go back to
Zinc binding site 2 out
of 2 in the Nucleocapsid Zinc Fingers Detected in Retroviruses: Exafs Studies on Intact Viruses and the Solution-State Structure of the Nucleocapsid Protein From Hiv-1
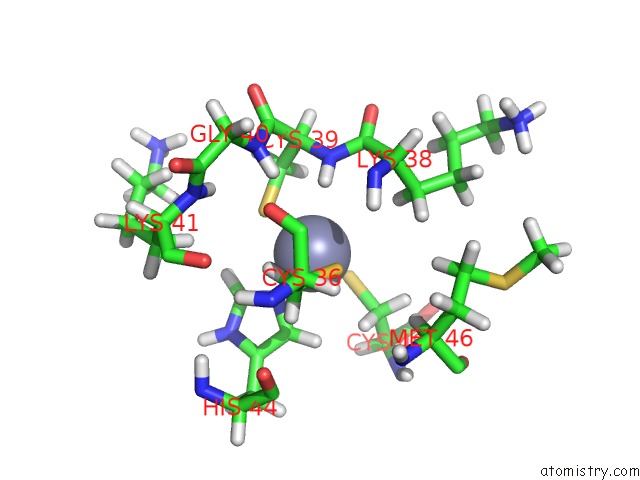
Mono view
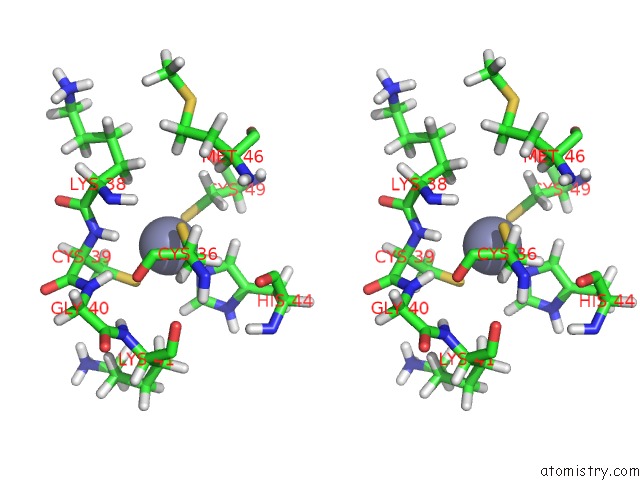
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Nucleocapsid Zinc Fingers Detected in Retroviruses: Exafs Studies on Intact Viruses and the Solution-State Structure of the Nucleocapsid Protein From Hiv-1 within 5.0Å range:
|
Reference:
M.F.Summers,
L.E.Henderson,
M.R.Chance,
J.W.Bess Jr.,
T.L.South,
P.R.Blake,
I.Sagi,
G.Perez-Alvarado,
R.C.Sowder 3Rd.,
D.R.Hare,
L.O.Arthur.
Nucleocapsid Zinc Fingers Detected in Retroviruses: Exafs Studies of Intact Viruses and the Solution-State Structure of the Nucleocapsid Protein From Hiv-1. Protein Sci. V. 1 563 1992.
ISSN: ISSN 0961-8368
PubMed: 1304355
Page generated: Sat Oct 12 21:55:15 2024
ISSN: ISSN 0961-8368
PubMed: 1304355
Last articles
Al in 7X24Al in 7X22
Al in 7X21
Al in 7X20
Al in 7W7W
Al in 7VSH
Al in 7VPL
Al in 7TUB
Al in 7VQ7
Al in 7VOJ