Zinc »
PDB 9qa6-9vxg »
9vck »
Zinc in PDB 9vck: Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex
Enzymatic activity of Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex
All present enzymatic activity of Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex:
2.1.1.56;
2.1.1.56;
Other elements in 9vck:
The structure of Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex also contains other interesting chemical elements:
| Calcium | (Ca) | 2 atoms |
| Fluorine | (F) | 1 atom |
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex
(pdb code 9vck). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 5 binding sites of Zinc where determined in the Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex, PDB code: 9vck:
Jump to Zinc binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Zinc where determined in the Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex, PDB code: 9vck:
Jump to Zinc binding site number: 1; 2; 3; 4; 5;
Zinc binding site 1 out of 5 in 9vck
Go back to
Zinc binding site 1 out
of 5 in the Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex
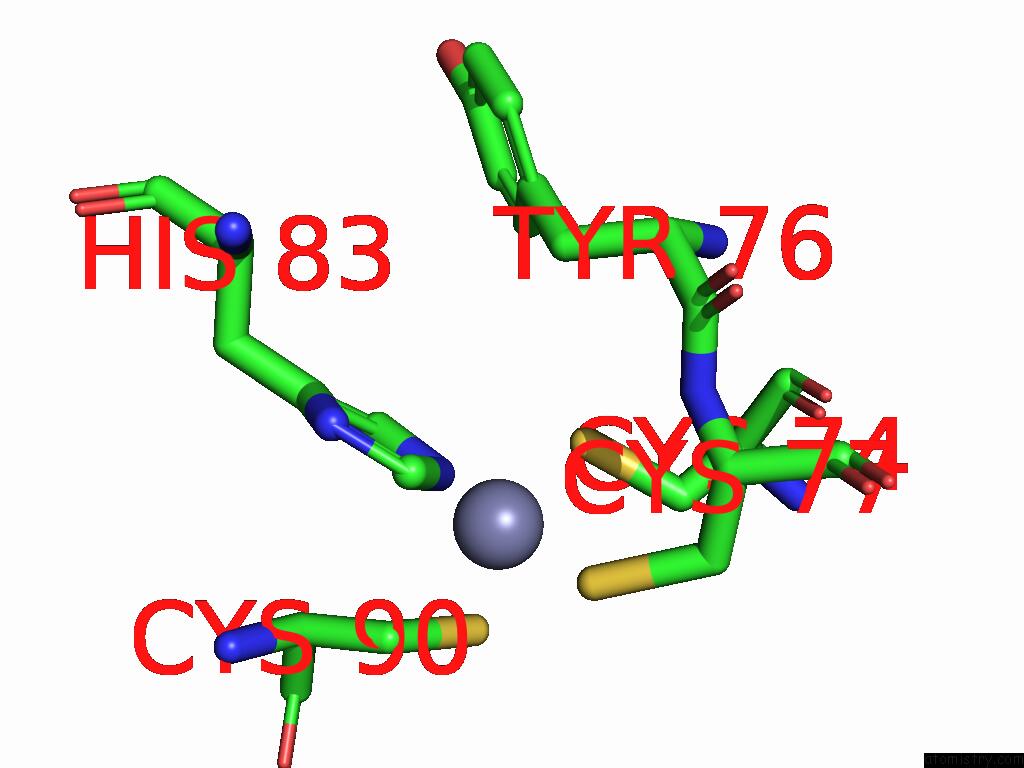
Mono view
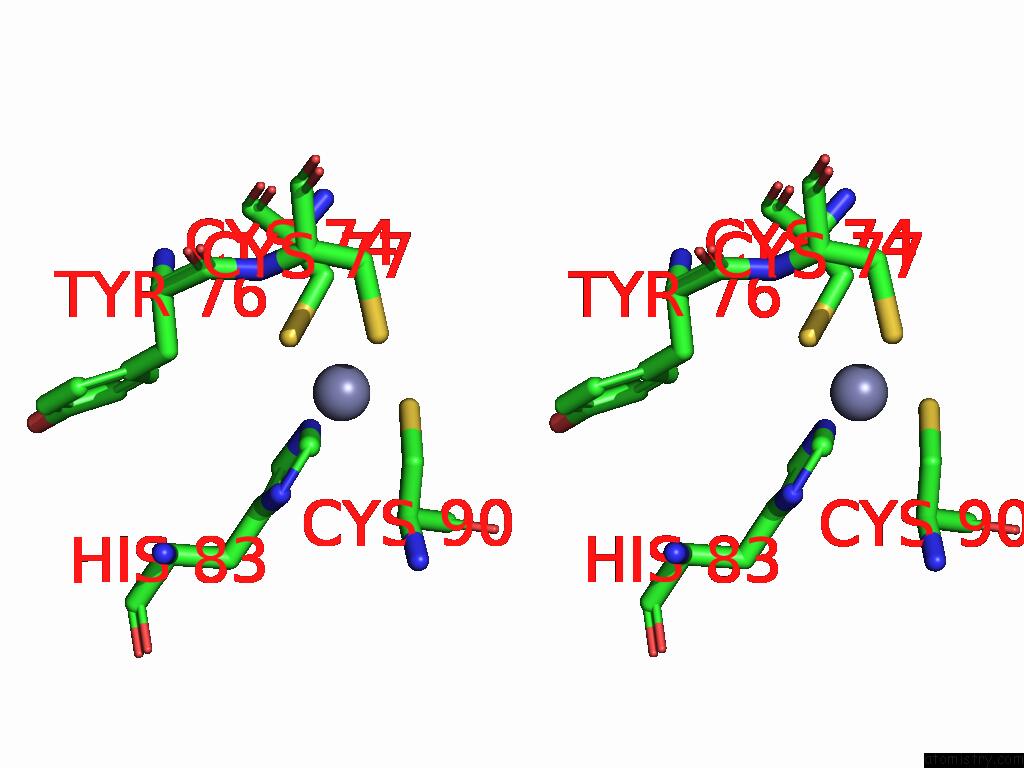
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex within 5.0Å range:
|
Zinc binding site 2 out of 5 in 9vck
Go back to
Zinc binding site 2 out
of 5 in the Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex
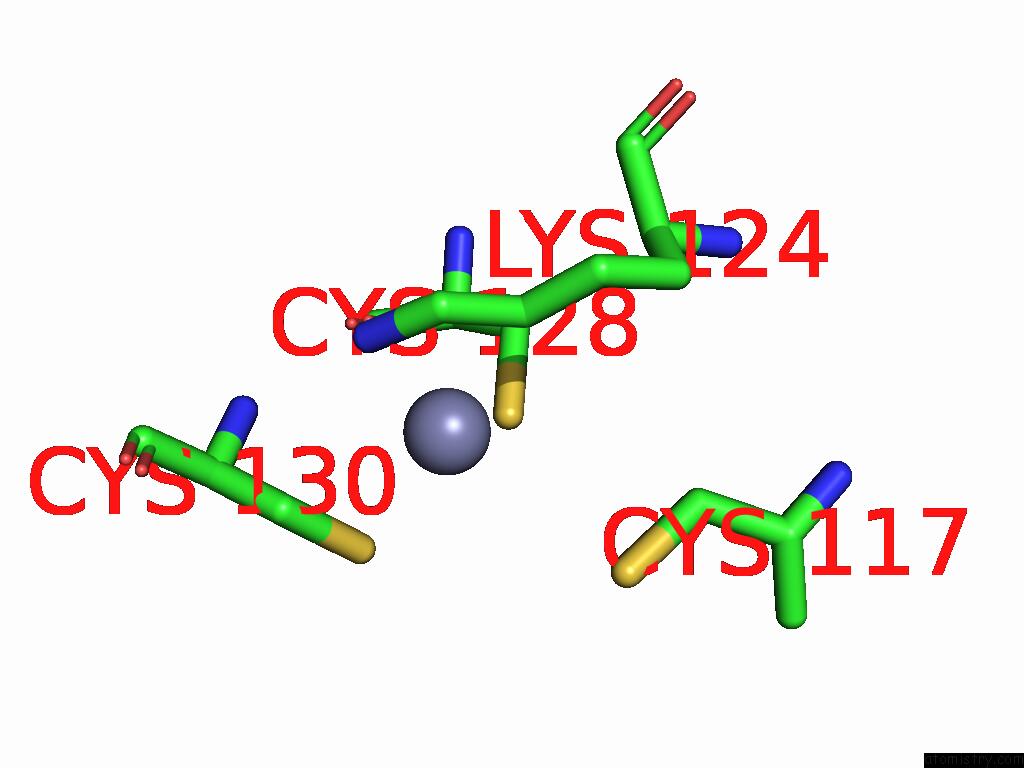
Mono view
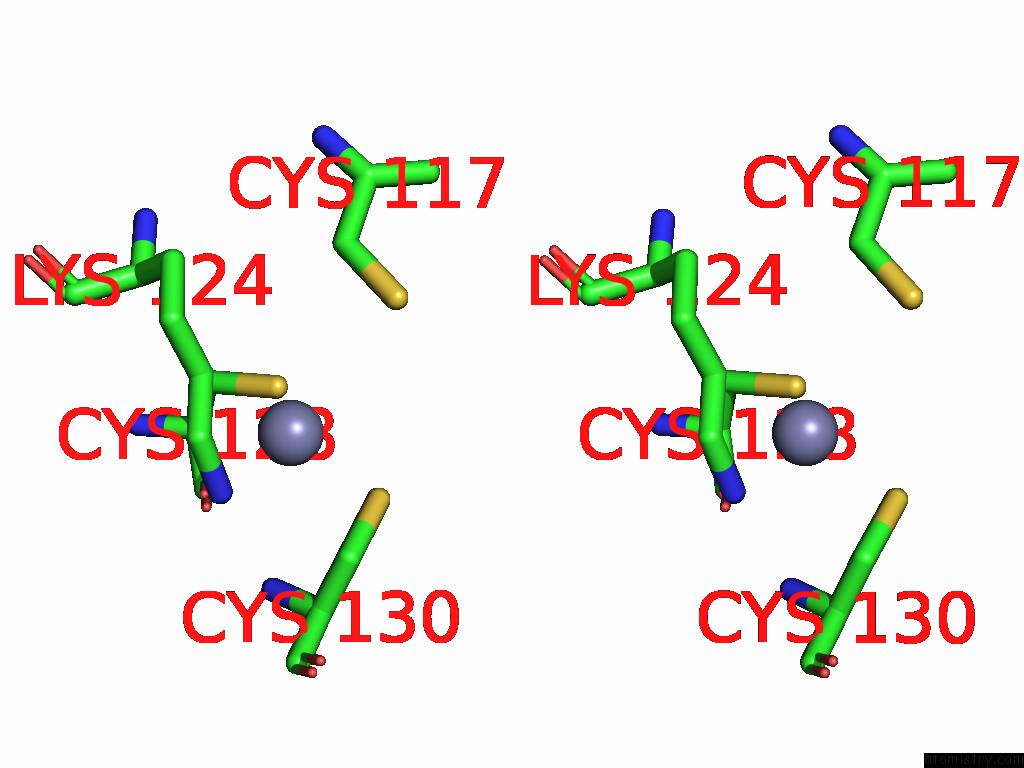
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex within 5.0Å range:
|
Zinc binding site 3 out of 5 in 9vck
Go back to
Zinc binding site 3 out
of 5 in the Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex
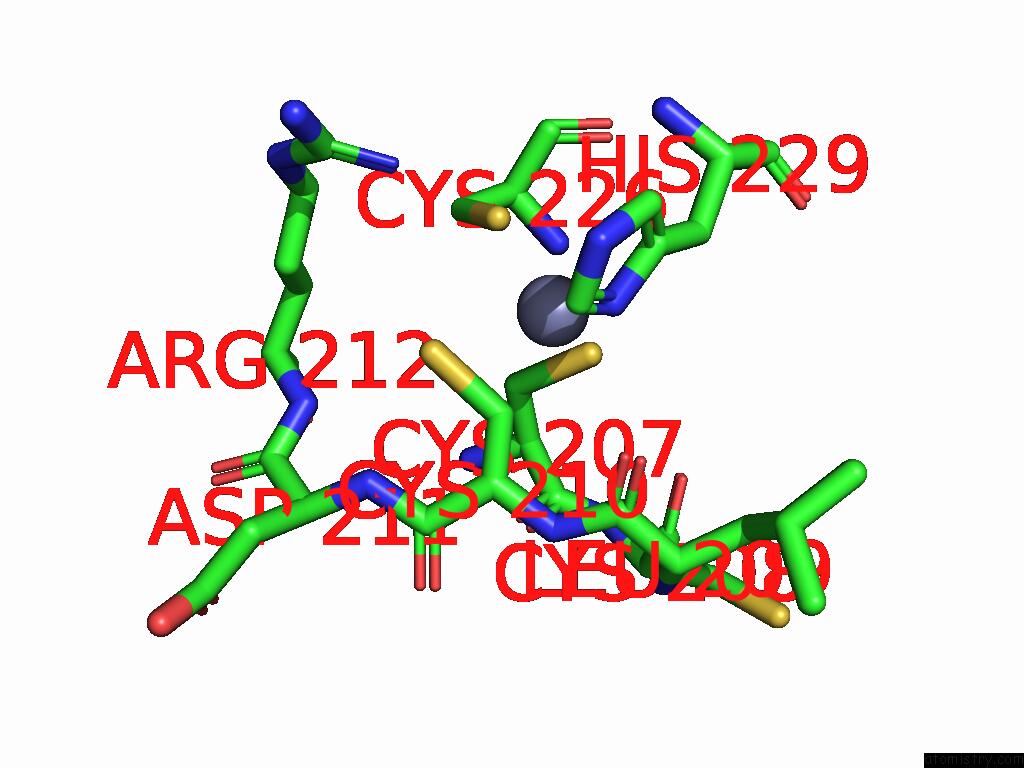
Mono view
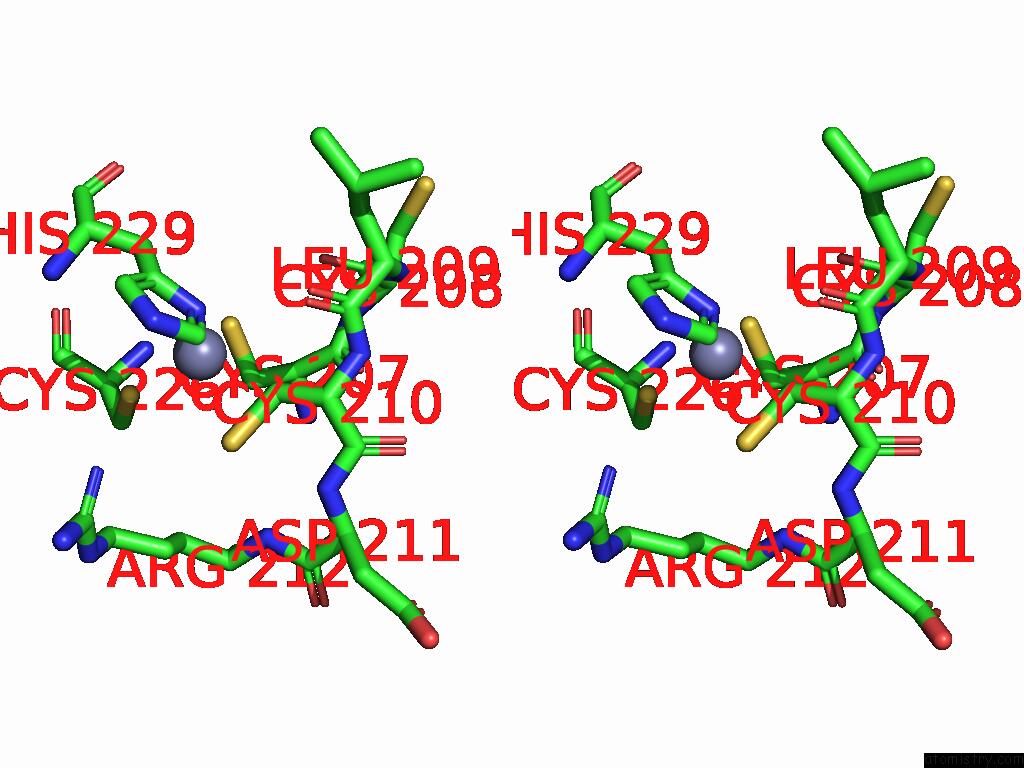
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex within 5.0Å range:
|
Zinc binding site 4 out of 5 in 9vck
Go back to
Zinc binding site 4 out
of 5 in the Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex
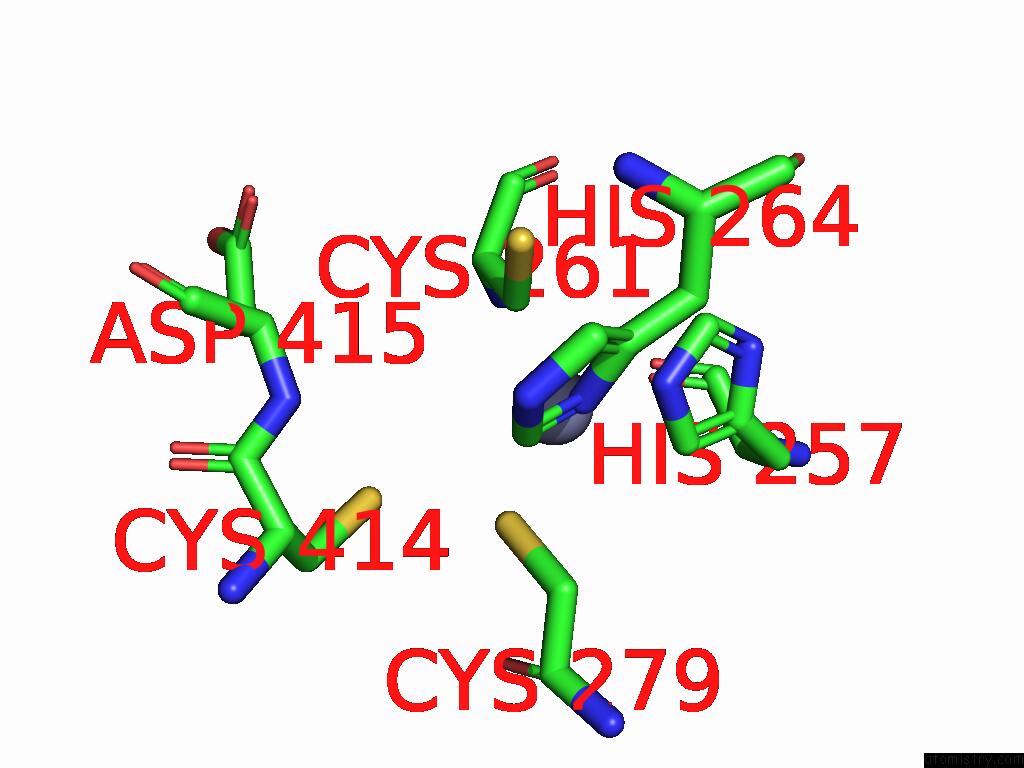
Mono view
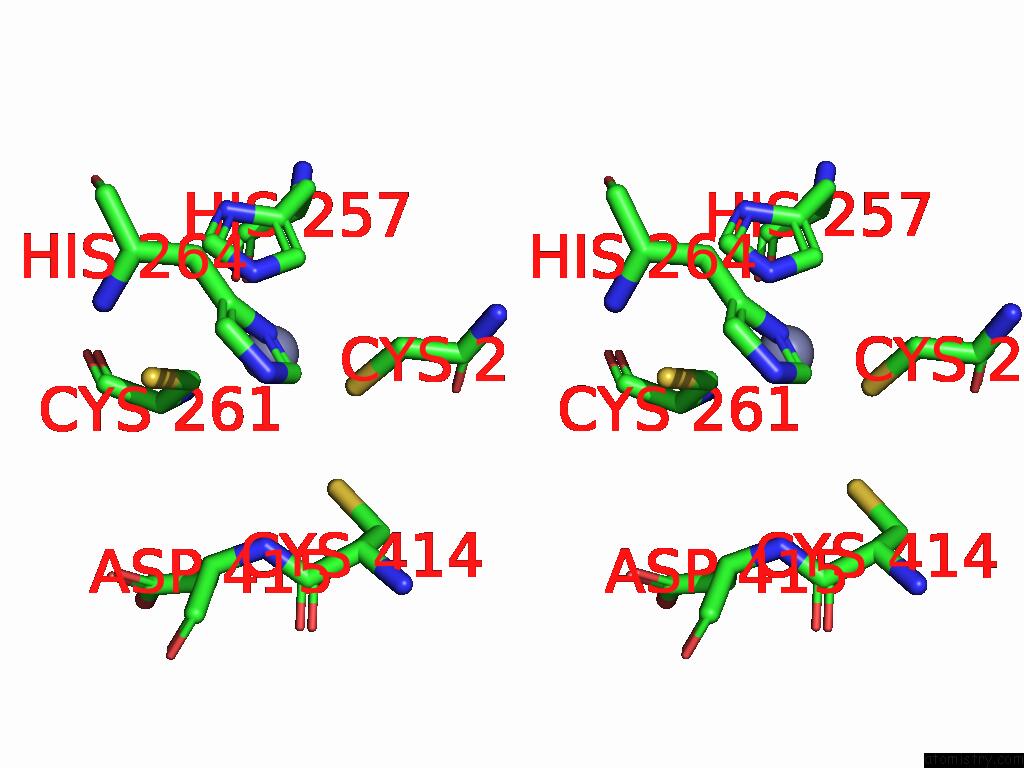
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 4 of Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex within 5.0Å range:
|
Zinc binding site 5 out of 5 in 9vck
Go back to
Zinc binding site 5 out
of 5 in the Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex
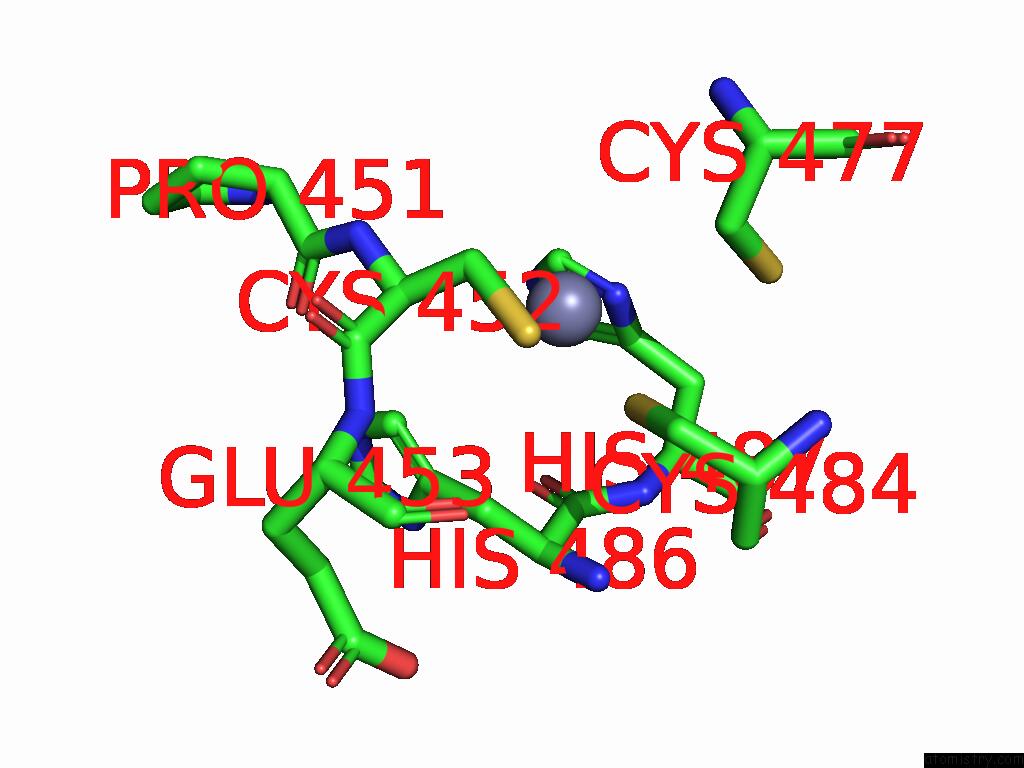
Mono view
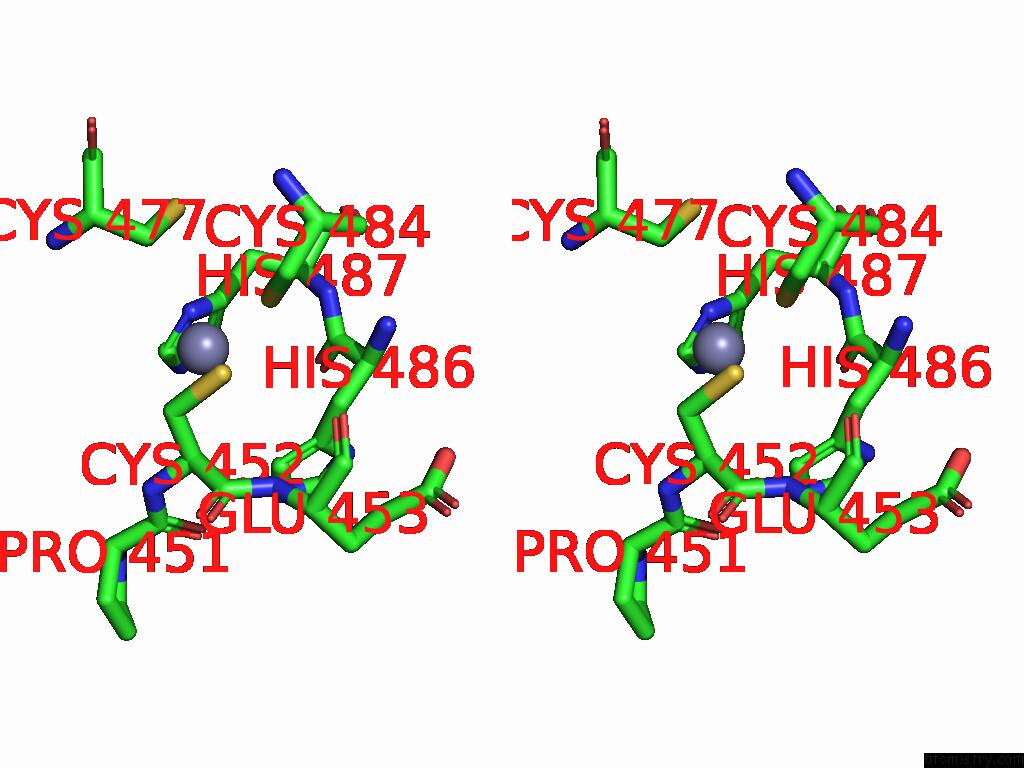
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 5 of Cryo-Em Structure of Sars-Cov-2 NSP10/NSP14:Rna:Smp Complex within 5.0Å range:
|
Reference:
J.Wang,
Y.Pan,
Y.Liu,
B.Huang,
G.Jin,
L.Zhang,
F.Zhou,
X.Chang,
Y.Huang,
L.Yan,
Y.Dong,
Z.Rao,
D.Liu,
Z.Lou.
Structural Basis and Rational Design of Nucleotide Analogue Inhibitor Evading the Sars-Cov-2 Proofreading Enzyme. J.Am.Chem.Soc. V. 147 21896 2025.
ISSN: ESSN 1520-5126
PubMed: 40509573
DOI: 10.1021/JACS.5C05014
Page generated: Fri Aug 22 18:59:33 2025
ISSN: ESSN 1520-5126
PubMed: 40509573
DOI: 10.1021/JACS.5C05014
Last articles
Zn in 9QM9Zn in 9S44
Zn in 9OFE
Zn in 9OFC
Zn in 9OFD
Zn in 9OF1
Zn in 9OFB
Zn in 9N0J
Zn in 9M5X
Zn in 9LGI