Zinc »
PDB 9ntg-9ueq »
9qlm »
Zinc in PDB 9qlm: Solution Structure of the TAF3-Phd Bound to A H3K4ME3Q5SER Histone Tail Peptide with A Serotonylated Glutamine
Zinc Binding Sites:
The binding sites of Zinc atom in the Solution Structure of the TAF3-Phd Bound to A H3K4ME3Q5SER Histone Tail Peptide with A Serotonylated Glutamine
(pdb code 9qlm). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the Solution Structure of the TAF3-Phd Bound to A H3K4ME3Q5SER Histone Tail Peptide with A Serotonylated Glutamine, PDB code: 9qlm:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the Solution Structure of the TAF3-Phd Bound to A H3K4ME3Q5SER Histone Tail Peptide with A Serotonylated Glutamine, PDB code: 9qlm:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 9qlm
Go back to
Zinc binding site 1 out
of 2 in the Solution Structure of the TAF3-Phd Bound to A H3K4ME3Q5SER Histone Tail Peptide with A Serotonylated Glutamine
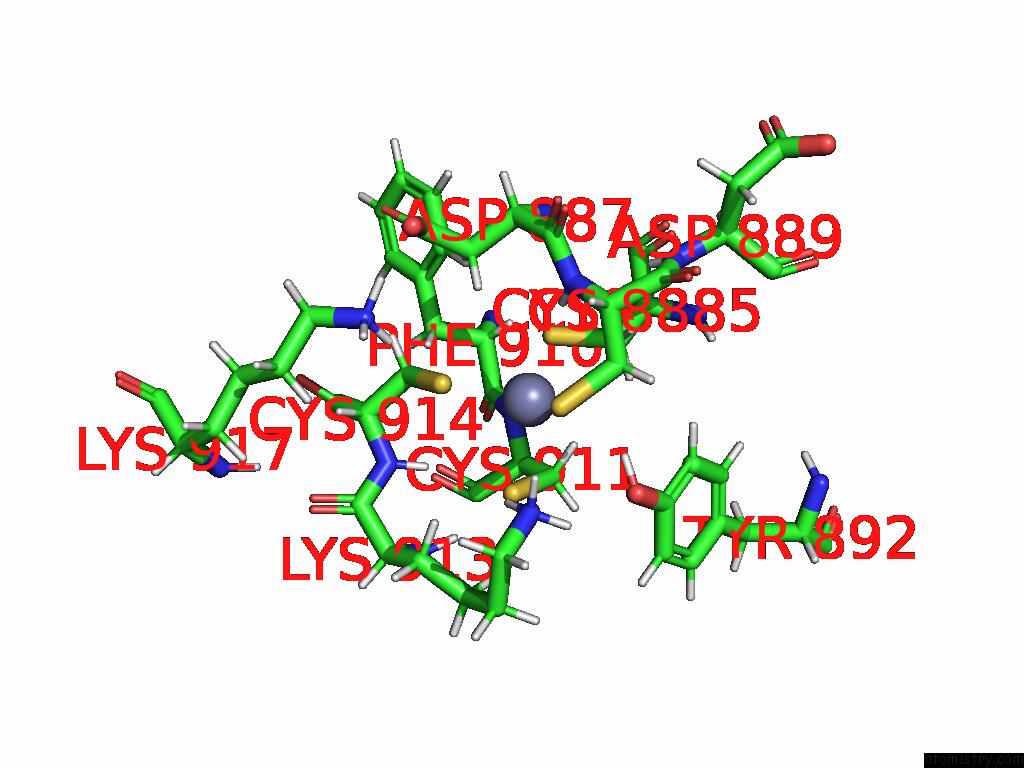
Mono view
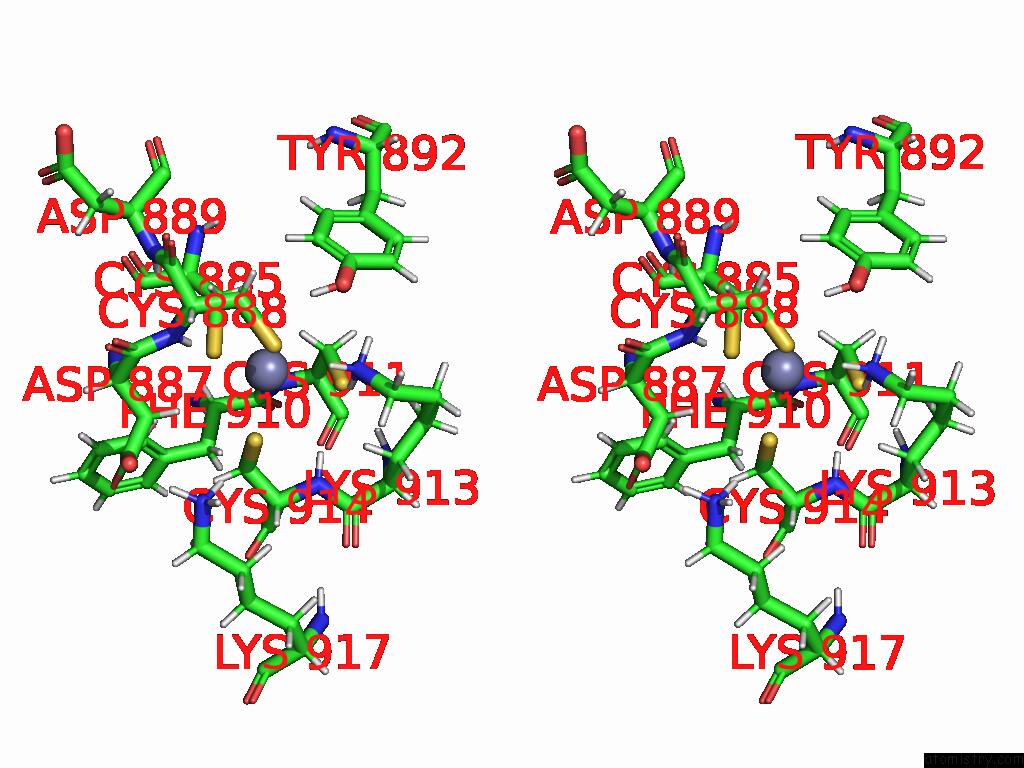
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Solution Structure of the TAF3-Phd Bound to A H3K4ME3Q5SER Histone Tail Peptide with A Serotonylated Glutamine within 5.0Å range:
|
Zinc binding site 2 out of 2 in 9qlm
Go back to
Zinc binding site 2 out
of 2 in the Solution Structure of the TAF3-Phd Bound to A H3K4ME3Q5SER Histone Tail Peptide with A Serotonylated Glutamine
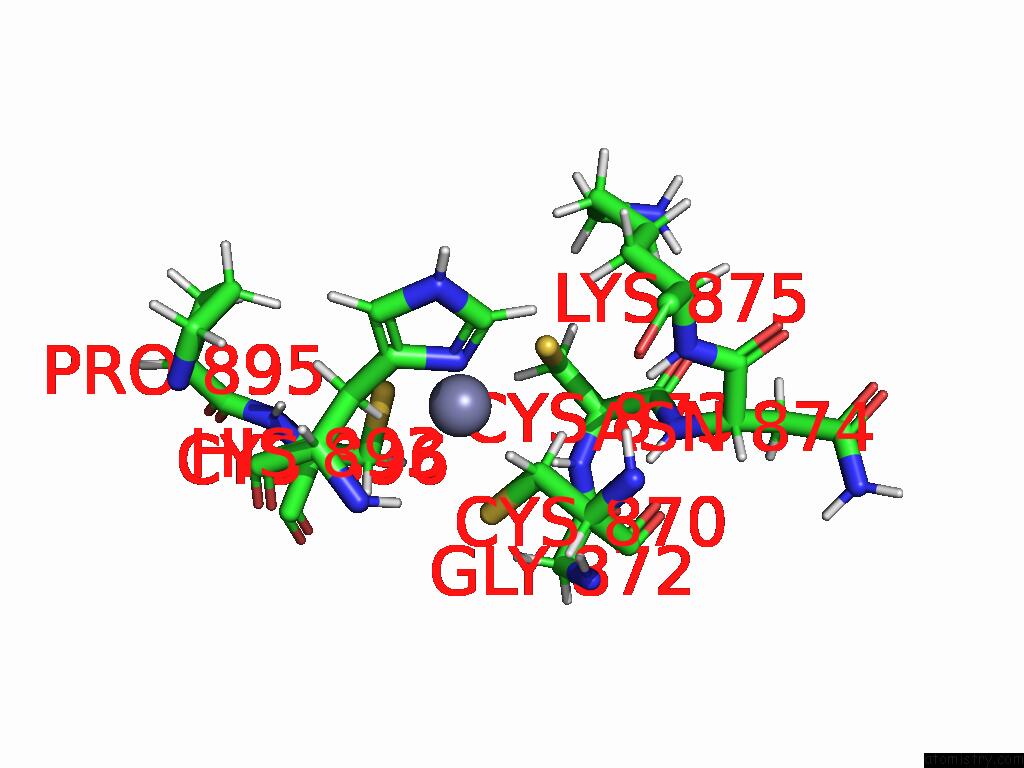
Mono view
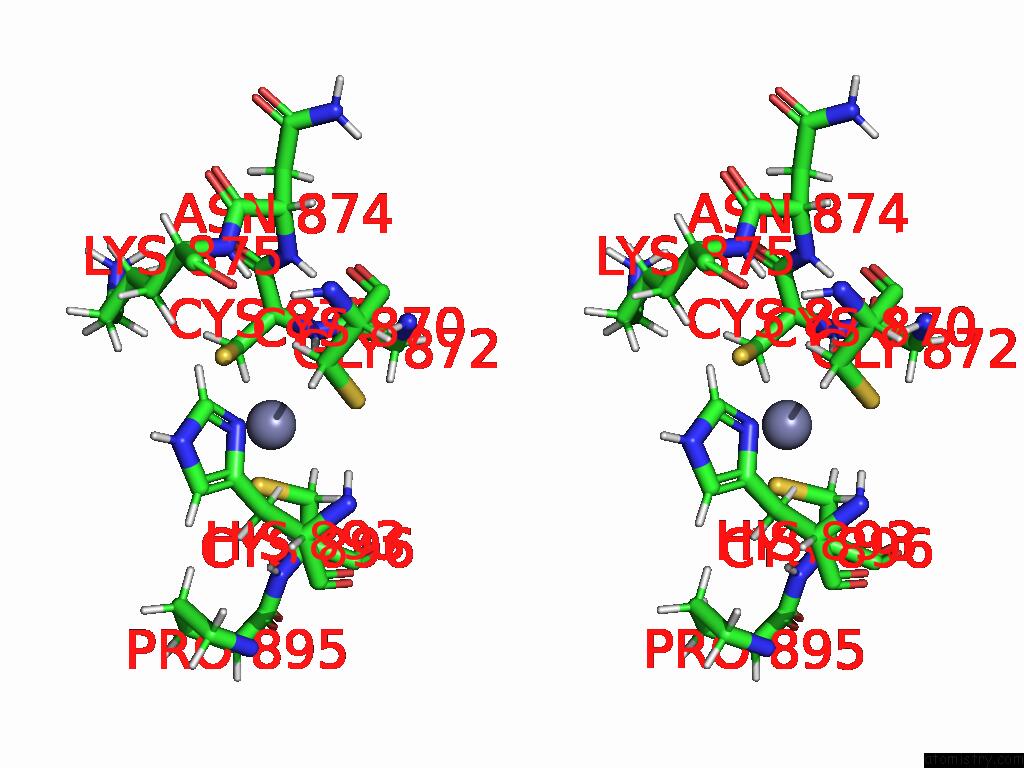
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Solution Structure of the TAF3-Phd Bound to A H3K4ME3Q5SER Histone Tail Peptide with A Serotonylated Glutamine within 5.0Å range:
|
Reference:
L.Pulido-Cortes,
H.Gielingh,
V.Thijssen,
M.Liu,
R.Yoshisada,
L.R.Soares,
N.Sheikh,
F.Friedrich,
H.Greschik,
L.Peng,
R.V.Honorato,
M.Jung,
A.M.J.J.Bonvin,
M.L.Biniossek,
R.Schule,
H.Van Ingen,
H.T.M.Timmers,
S.Jongkees.
Molecular Determinants For Recognition of Serotonylated Chromatin To Be Published.
Page generated: Fri Aug 22 18:57:07 2025
Last articles
Ca in 9K6MCa in 9IUJ
Ca in 9JD0
Ca in 9JD1
Ca in 9JCX
Ca in 9I6C
Ca in 9GTB
Ca in 9GSH
Ca in 9GSI
Ca in 9GRG