Zinc »
PDB 9ntg-9ueq »
9qa6 »
Zinc in PDB 9qa6: Cryo-Em Structure of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (S32-N876)
Zinc Binding Sites:
The binding sites of Zinc atom in the Cryo-Em Structure of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (S32-N876)
(pdb code 9qa6). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 3 binding sites of Zinc where determined in the Cryo-Em Structure of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (S32-N876), PDB code: 9qa6:
Jump to Zinc binding site number: 1; 2; 3;
In total 3 binding sites of Zinc where determined in the Cryo-Em Structure of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (S32-N876), PDB code: 9qa6:
Jump to Zinc binding site number: 1; 2; 3;
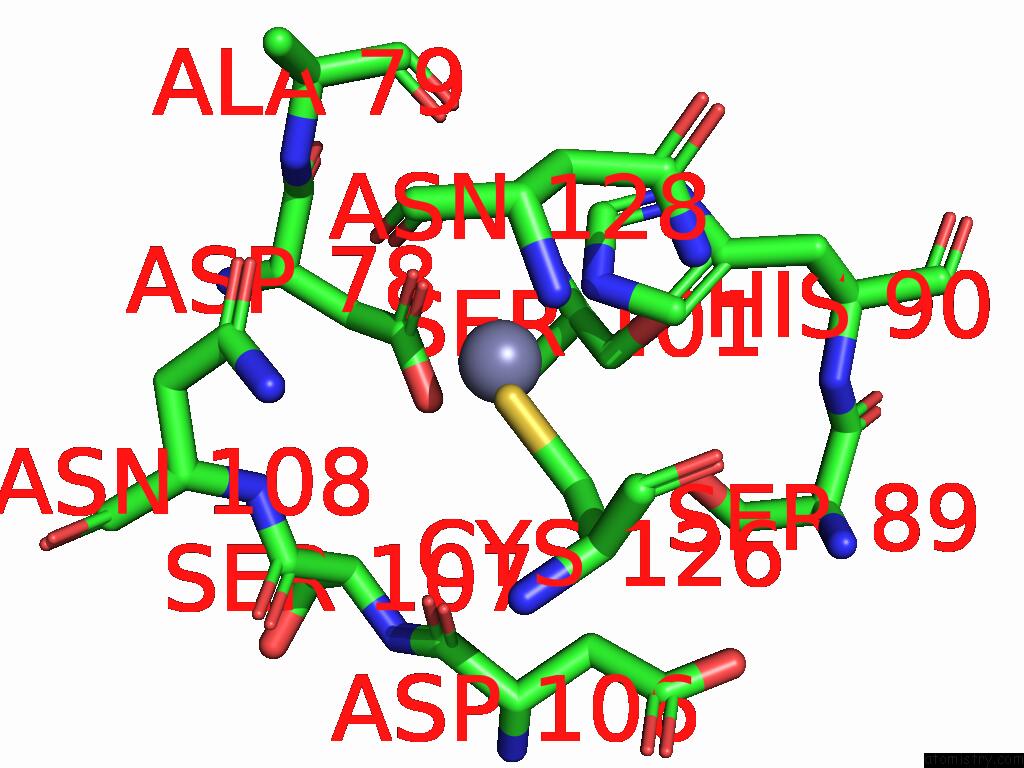
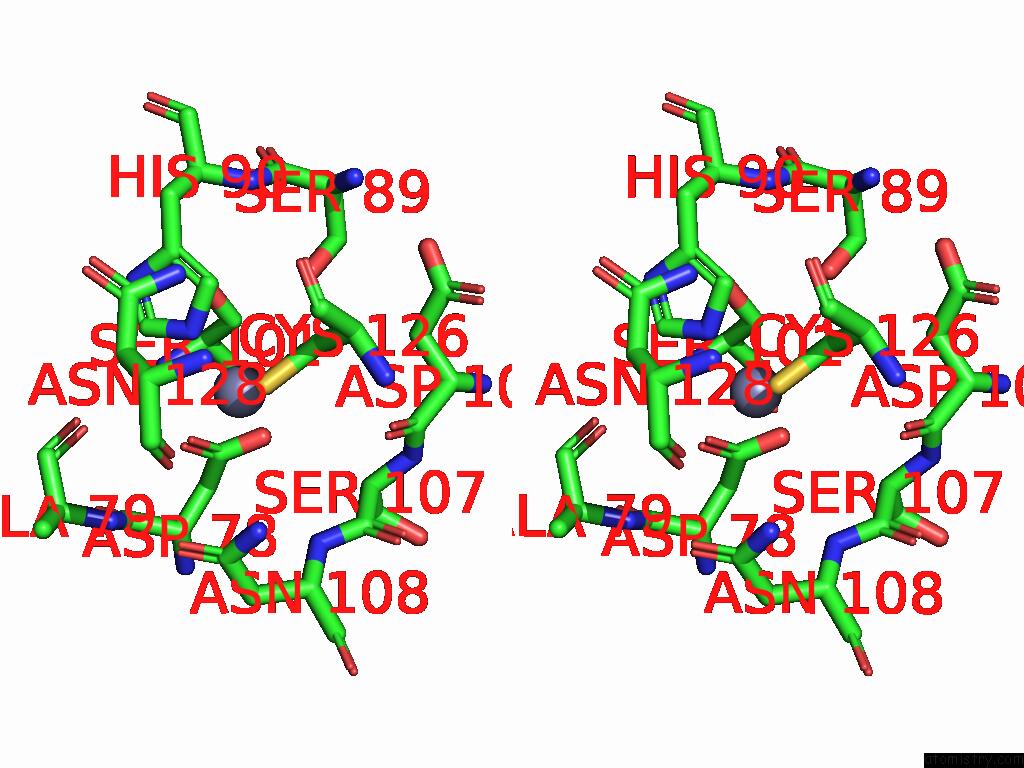
Zinc binding site 1 out of 3 in 9qa6
Go back to
Zinc binding site 1 out
of 3 in the Cryo-Em Structure of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (S32-N876)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of Cryo-Em Structure of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (S32-N876) within 5.0Å range:
|
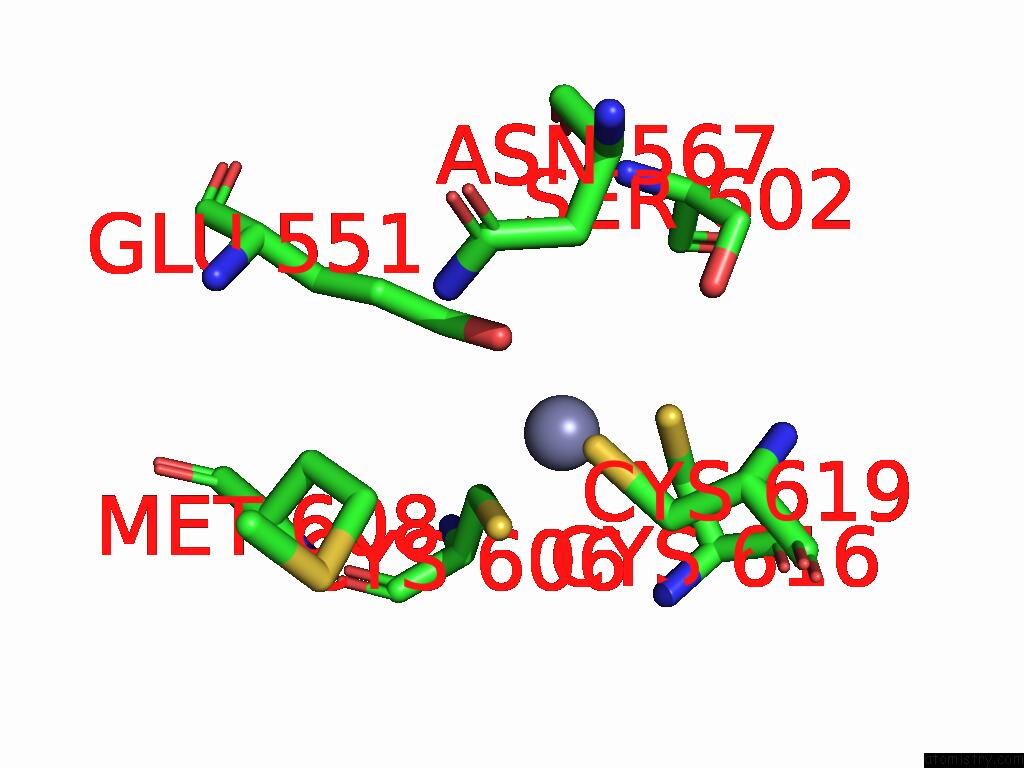
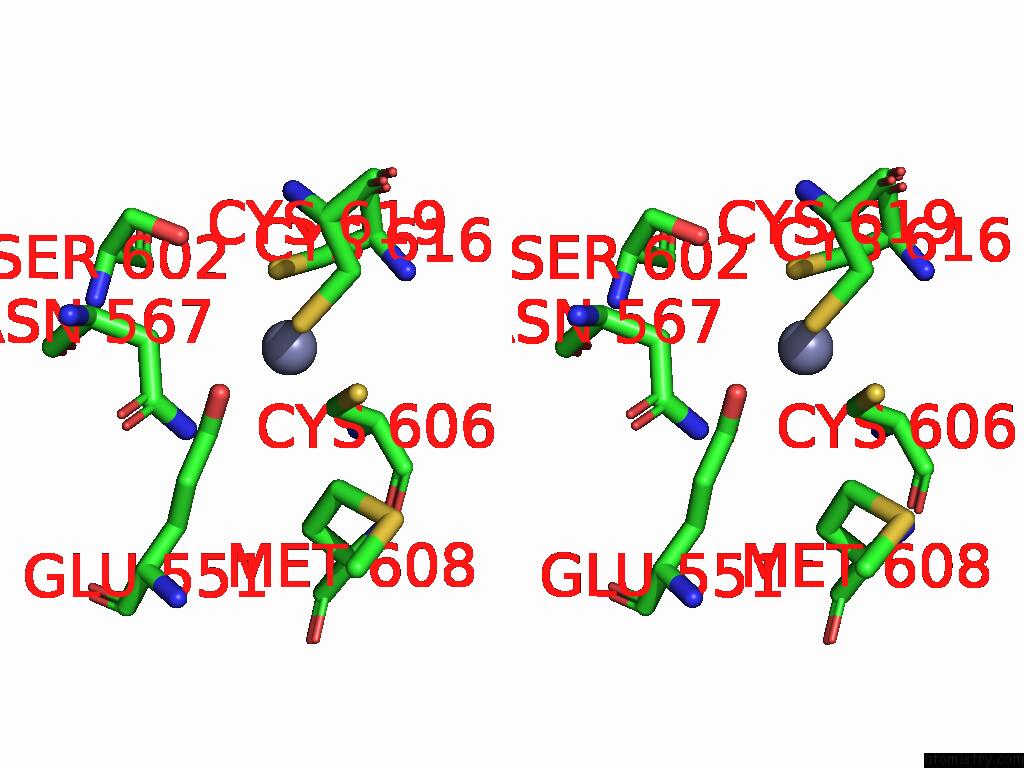
Zinc binding site 2 out of 3 in 9qa6
Go back to
Zinc binding site 2 out
of 3 in the Cryo-Em Structure of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (S32-N876)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of Cryo-Em Structure of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (S32-N876) within 5.0Å range:
|
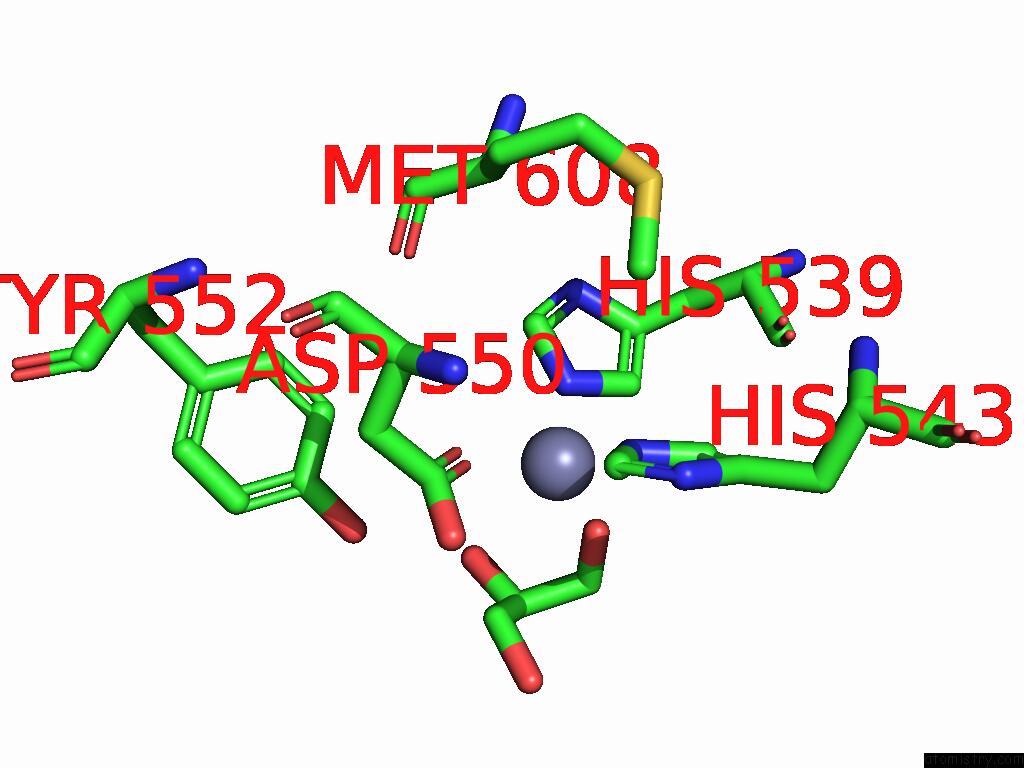
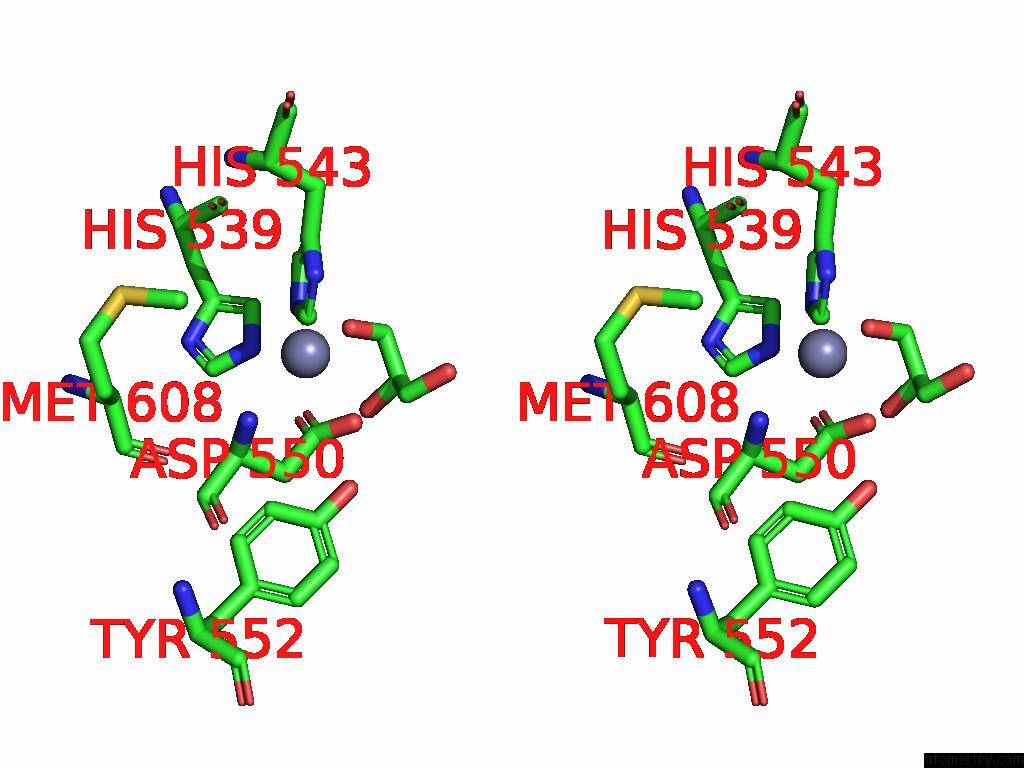
Zinc binding site 3 out of 3 in 9qa6
Go back to
Zinc binding site 3 out
of 3 in the Cryo-Em Structure of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (S32-N876)
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 3 of Cryo-Em Structure of Thomasclavelia Ramosa Iga Peptidase (Igase) Active Site Mutant (S32-N876) within 5.0Å range:
|
Reference:
J.S.Ramirez-Larrota,
U.Eckhard,
P.Guerra,
P.Juyoux,
F.X.Gomis-Ruth.
Thomasclavelia Ramosa Iga Peptidase Active Site Mutant - E540A (Including Ntd) To Be Published.
Page generated: Fri Aug 22 18:54:16 2025
Last articles
Ca in 9K6MCa in 9IUJ
Ca in 9JD0
Ca in 9JD1
Ca in 9JCX
Ca in 9I6C
Ca in 9GTB
Ca in 9GSH
Ca in 9GSI
Ca in 9GRG