Zinc »
PDB 9lxo-9ntf »
9mse »
Zinc in PDB 9mse: De Novo Sign Rna Polymerase Transcription Initiation Intermediate with Pre-Catalytic Bebp State (RPI1 Open Ring)
Enzymatic activity of De Novo Sign Rna Polymerase Transcription Initiation Intermediate with Pre-Catalytic Bebp State (RPI1 Open Ring)
All present enzymatic activity of De Novo Sign Rna Polymerase Transcription Initiation Intermediate with Pre-Catalytic Bebp State (RPI1 Open Ring):
2.7.7.6;
2.7.7.6;
Other elements in 9mse:
The structure of De Novo Sign Rna Polymerase Transcription Initiation Intermediate with Pre-Catalytic Bebp State (RPI1 Open Ring) also contains other interesting chemical elements:
| Magnesium | (Mg) | 3 atoms |
Zinc Binding Sites:
The binding sites of Zinc atom in the De Novo Sign Rna Polymerase Transcription Initiation Intermediate with Pre-Catalytic Bebp State (RPI1 Open Ring)
(pdb code 9mse). This binding sites where shown within
5.0 Angstroms radius around Zinc atom.
In total 2 binding sites of Zinc where determined in the De Novo Sign Rna Polymerase Transcription Initiation Intermediate with Pre-Catalytic Bebp State (RPI1 Open Ring), PDB code: 9mse:
Jump to Zinc binding site number: 1; 2;
In total 2 binding sites of Zinc where determined in the De Novo Sign Rna Polymerase Transcription Initiation Intermediate with Pre-Catalytic Bebp State (RPI1 Open Ring), PDB code: 9mse:
Jump to Zinc binding site number: 1; 2;
Zinc binding site 1 out of 2 in 9mse
Go back to
Zinc binding site 1 out
of 2 in the De Novo Sign Rna Polymerase Transcription Initiation Intermediate with Pre-Catalytic Bebp State (RPI1 Open Ring)
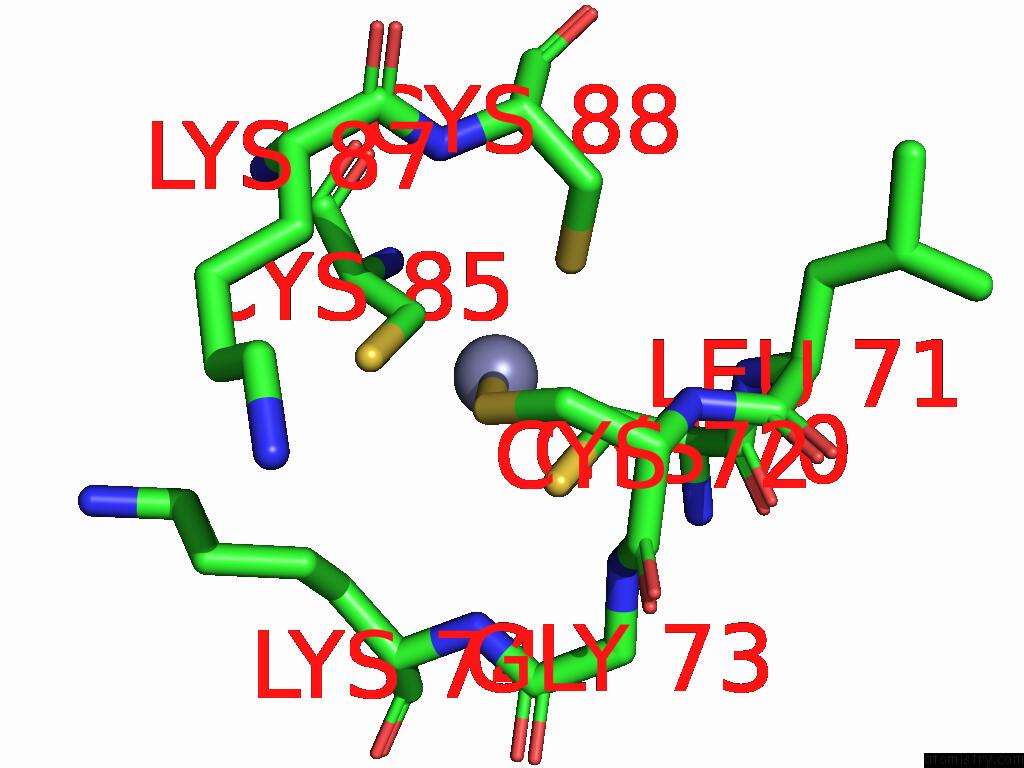
Mono view
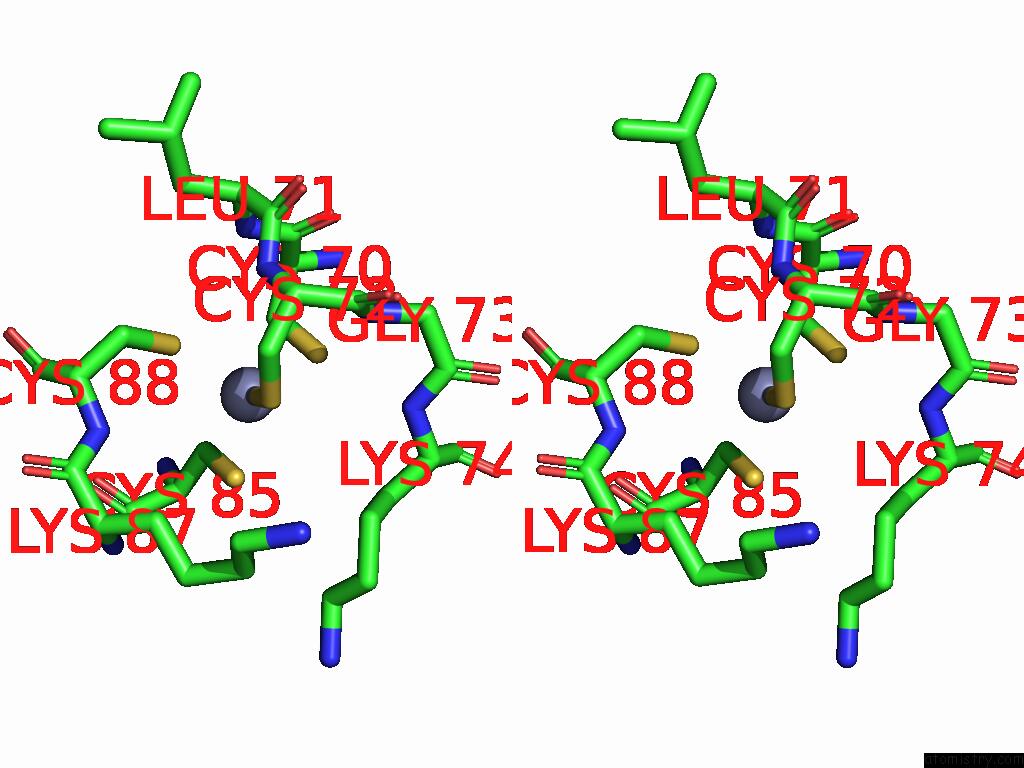
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 1 of De Novo Sign Rna Polymerase Transcription Initiation Intermediate with Pre-Catalytic Bebp State (RPI1 Open Ring) within 5.0Å range:
|
Zinc binding site 2 out of 2 in 9mse
Go back to
Zinc binding site 2 out
of 2 in the De Novo Sign Rna Polymerase Transcription Initiation Intermediate with Pre-Catalytic Bebp State (RPI1 Open Ring)
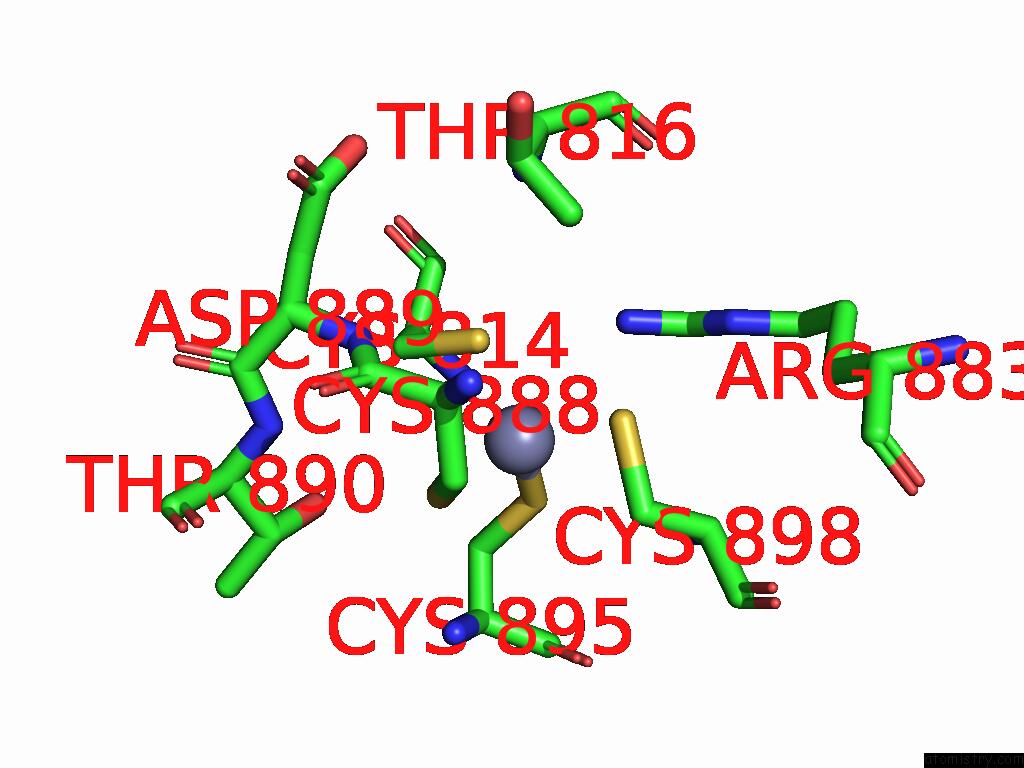
Mono view
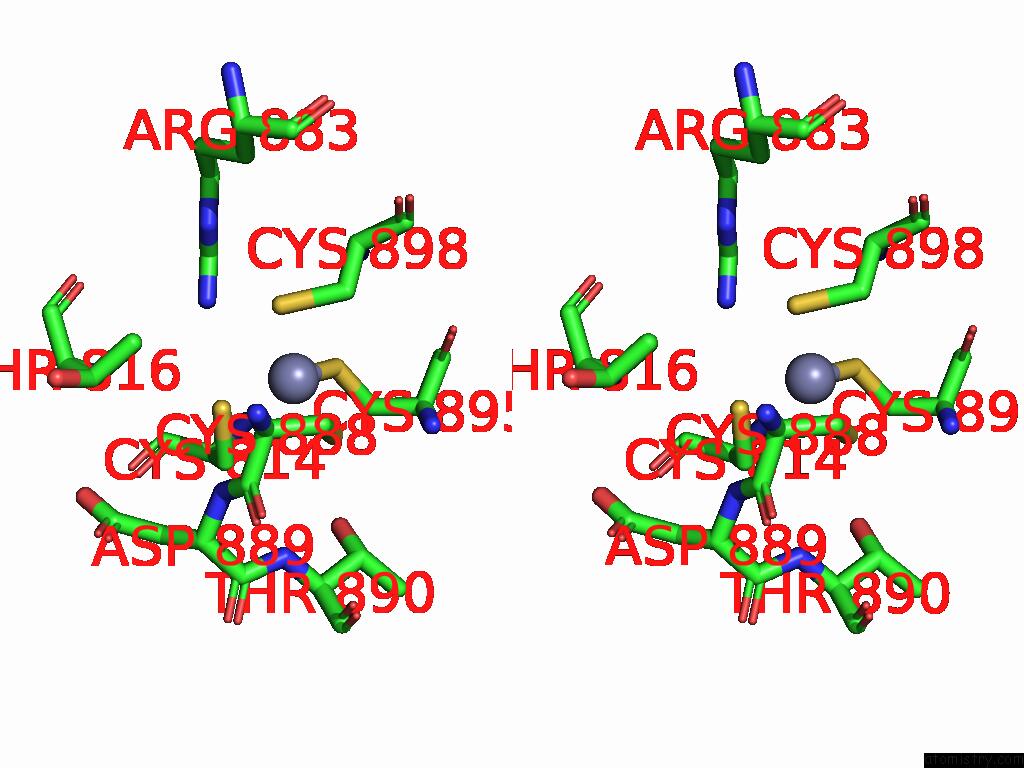
Stereo pair view

Mono view

Stereo pair view
A full contact list of Zinc with other atoms in the Zn binding
site number 2 of De Novo Sign Rna Polymerase Transcription Initiation Intermediate with Pre-Catalytic Bebp State (RPI1 Open Ring) within 5.0Å range:
|
Reference:
A.U.Mueller,
N.Molina,
B.T.Nixon,
S.A.Darst.
Real-Time Capture of Sigma N Transcription Initiation Intermediates Reveals Mechanism of Atpase-Driven Activation By Limited Unfolding. Nat Commun V. 16 7138 2025.
ISSN: ESSN 2041-1723
PubMed: 40759887
DOI: 10.1038/S41467-025-61837-4
Page generated: Fri Aug 22 18:42:34 2025
ISSN: ESSN 2041-1723
PubMed: 40759887
DOI: 10.1038/S41467-025-61837-4
Last articles
Ca in 9GCYCa in 9G8H
Ca in 9G8V
Ca in 9EJF
Ca in 9EJE
Ca in 9EIT
Ca in 9CYJ
Ca in 9DBB
Ca in 9DB8
Ca in 9CYE